Abstract
Streptomyces genomes encode two homologs of the nucleoid-associated HU proteins. One of them, here designated HupA, is of a conventional type similar to E. coli HUα and HUβ, while the other, HupS, is a two-domain protein. In addition to the N-terminal part that is similar to that of HU proteins, it has a C-terminal domain that is similar to the alanine- and lysine-rich C termini of eukaryotic linker histones. Such two-domain HU proteins are found only among Actinobacteria. In this phylum some organisms have only a single HU protein of the type with a C-terminal histone H1-like domain (e.g., Hlp in Mycobacterium smegmatis), while others have only a single conventional HU. Yet others, including the streptomycetes, produce both types of HU proteins. We show here that the two HU genes in Streptomyces coelicolor are differentially regulated and that hupS is specifically expressed during sporulation, while hupA is expressed in vegetative hyphae. The developmental upregulation of hupS occurred in sporogenic aerial hyphal compartments and was dependent on the developmental regulators whiA, whiG, and whiI. HupS was found to be nucleoid associated in spores, and a hupS deletion mutant had an average nucleoid size in spores larger than that in the parent strain. The mutant spores were also defective in heat resistance and spore pigmentation, although they possessed apparently normal spore walls and displayed no increased sensitivity to detergents. Overall, the results show that HupS is specifically involved in sporulation and may affect nucleoid architecture and protection in spores of S. coelicolor.
Bacteria face the formidable task of compacting their chromosomes to accommodate them in a small cytoplasmic volume and at the same time maintaining the nucleoids in a highly organized and dynamic state so that transcription, DNA replication, and chromosome partitioning can take place with accuracy and speed. DNA also has to be protected from damage to preserve the genomic information, for example, during periods of nongrowth. The structure and organization of bacterial chromatin are shaped by compacting forces, like DNA supercoiling and macromolecular crowding, and by small, basic, nucleoid-associated proteins (reviewed in, e.g., references 33, 34, 50, and 54). Such proteins are often referred to as histone like, although they share no sequence homology with eukaryotic histones and constitute a very heterogeneous group. They include HU-like proteins, which are ubiquitous in bacteria and often abundant, for example, Escherichia coli HU, which is a homo- or heterodimer of the homologous subunits α and β (21). HU binds to DNA without sequence specificity and can contribute to nucleoid compaction by bending or wrapping DNA. Other nucleoid-associated proteins, like IHF, H-NS, Lrp, and Fis, can wrap or bridge segments of DNA (33, 34). Together with the condensin/cohesin-like SMC and MukB proteins, they contribute to the global compaction and dynamic organization of bacterial chromosomes, as well as to local effects on DNA topology that affect the expression of specific genes.
Under conditions of nongrowth and long-term survival, chromatin structure changes and additional proteins are produced that are specifically involved in packaging and protecting the DNA. For example, the relative amounts of the different nucleoid-associated proteins in E. coli change when cells enter stationary phase (4). Particularly, Dps accumulates to high levels in stationary-phase E. coli cells and forms large crystal-like nucleoprotein complexes with DNA (2, 19). Dps helps to protect against oxidative stress, heat, and UV irradiation (2, 40). Other well-known examples are the α/β-type small acid-soluble spore proteins (SASPs), which form tight nucleoprotein complexes with endospore DNA in Bacillus and some other gram-positive genera (32, 48). The SASPs are a main factor contributing to the extreme resistance and ability to survive adverse conditions that are displayed by endospores.
Bacteria of the genus Streptomyces, which belong to the phylum Actinobacteria, also produce spores (9, 16). These spores are fundamentally different from the endospores of Firmicutes and are formed by division of the long apical cell of a hypha into a chain of spores. The Streptomyces spores lack, among other things, the surface layers that are contributed from the mother cell in endospores and are significantly less resistant to adverse conditions compared to Bacillus endospores (14). Yet, spore formation is a crucial part of the Streptomyces life cycle, and streptomycetes may be present mostly as spores in the soil environment (14). Thus, the ability of Streptomyces spores to preserve their DNA and survive for long periods of time is important, but the mechanisms for this are different from those found in, e.g., Bacillus. Streptomyces spores do not contain Ca2+-dipicolinate or SASPs (5, 14), which are crucial for mounting resistance in Bacillus species (48). Despite this, the spore DNA is condensed, and the spores show some degree of resistance to, for example, heat, desiccation, and sonication, compared to vegetative cells. The molecular and genetic basis for DNA packaging and protection in Streptomyces remains poorly understood.
Vegetatively, streptomycetes grow as hyphae and form mycelial networks (15). The sporulation pathway has been investigated mostly with Streptomyces coelicolor. In this organism, an aerial mycelium emerges on the surface of developing colonies, and the spores are formed by these specialized aerial hyphae. This developmental process is controlled by a complex regulatory network that is only partially understood (recently reviewed in references 13 and 16). The metamorphosis of the apical aerial hyphal cells to spores involves developmentally regulated changes in patterns of cell wall growth, cell division, and chromosome segregation. It requires, for example, the upregulation of cell division gene ftsZ and chromosome partitioning genes parA and parB, and this depends on regulatory genes like whiA, whiB, whiG, whiH, and whiI (18, 23). As a result, the long and multigenomic hyphal cell is divided into prespore compartments by multiple synchronized cell division events, each prespore simultaneously receiving a single chromosome. During the subsequent maturation of the spores, a thick spore wall is assembled and the spores are rounded up to an ovoid shape. Before release of individual spores from the spore chain, they acquire a gray spore pigment, which is biosynthetically related to polyketide antibiotics and specified by genes in the whiE gene cluster (27).
Several mutants are known that produce spores with poor resistance to adverse conditions. The actin-like MreB protein is required for normal assembly of the spore wall, and mreB mutant spores are significantly less resistant to heat and detergent compared to wild-type spores (36). Also mutants lacking whiD, which encodes a putative regulatory protein of a class that is unique to Actinobacteria, produce spores that are unusually heat sensitive and also show deficiencies and irregularities in formation of the spore wall (37). Furthermore, sigF mutants have spore walls that are thinner than normal ones, appear to have less compact nucleoids, and show increased sensitivity to detergent compared to the parent strain (43). The sigF gene encodes an RNA polymerase σ factor that is specifically expressed in sporulating aerial hyphae (28), but no sigF-dependent genes have yet been identified, except for whiE ORFVIII, which is required for processing of the gray spore pigment (27). Defects in spore wall structure coinciding with less compact nucleoids and increased heat sensitivity have also been reported for an smeA-sffA mutant, and most of this phenotype was ascribed to smeA, which encodes a small putative membrane protein of unknown function (3). In all these studies, the observed phenotypes were pleiotropic, and it was not possible to separate the decreased resistance or fitness of the spores from observable effects on the cell wall structure. It is therefore unclear what role nucleoid compaction may have in mounting resistance of the spore to adverse conditions and how the compaction is brought about.
Recently, it was reported that an smc mutant of S. coelicolor has less condensed nucleoids than its parent during early stages of sporulation (around the time when sporulation septa are formed) (30), but a parallel study did not detect any effects on nucleoid size or condensation in mature spores (12). This is consistent with the appearance of SMC foci on nucleoids during the early stages of spore formation and their disappearance during later stages (30). Thus, SMC seems to affect the nucleoid structure and segregation during formation of the prespores but may not directly affect packaging of chromosomes in mature spores.
Since relatively few genes that are directly involved in development of spores in S. coelicolor have so far been identified, we carried out a transcriptomic survey of gene expression during sporulation. One of the genes found to be developmentally regulated in this study encodes an HU-like protein with an additional C-terminal domain that is related to eukaryotic histone H1. As reported previously (53) and confirmed by genome sequencing (5), S. coelicolor carries two genes for HU-like proteins. We report here that these two genes are differentially regulated, with hupA (SCO2950, previously designated hup) being preferentially expressed in vegetative hyphae and with hupS (SCO5556, previously designated hup2) being developmentally controlled and strongly upregulated in sporogenic aerial hyphae. It is also shown that HupS is associated with spore nucleoids, that hupS is required for mounting full heat resistance of spores, and that it influences the condensation of nucleoids in mature spores. Thus, HupS is specifically involved in sporulation, and the S. coelicolor HupA and HupS proteins appear to have specialized roles in nucleoid architecture during different developmental stages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids that were used in this study are shown in Table 1. E. coli strain DH5α was the host for plasmid construction, and strain ET12567/pUZ8002 was used as a donor for conjugative transfer of nonmethylated plasmid DNA to S. coelicolor A3(2) strains. E. coli strain GM2929 was used to propagate nonmethylated plasmid DNA for transformation to Streptomyces protoplasts. S. coelicolor A3(2) strain M145 and its derivates were grown at 30°C. Media used for E. coli strains were Difco nutrient agar and broth if hygromycin was used for selection and Luria-Bertani media for other antibiotics. S. coelicolor strains were grown on mannitol soya flour (MS) agar or in yeast extract-malt extract medium (29). Antibiotics were used when appropriate at the following concentrations: apramycin at 25 μg/ml, hygromycin at 25 μg/ml, and kanamycin at 5 μg/ml for S. coelicolor and apramycin at 50 μg/ml, carbenicillin at 50 μg/ml, hygromycin at 100 μg/ml, kanamycin at 50 μg/ml, and nalidixic acid at 20 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| S. coelicolor A3(2) strains | ||
| J2400 | whiG::hyg | 17 |
| J2401 | whiA::hyg | 17 |
| J2408 | ΔwhiH::ermE | 17 |
| J2450 | whiI::hyg | 4 |
| K304 | ΔhupS::Ωhyg | This work |
| K306 | hupA::pKF290[hupA-egfp] | This work |
| K307 | hupS::pKF292[hupS-egfp] | This work |
| K308 | hupS::pKF293[hupS-egfp] | This work |
| K309 | whiA::hyg hupS::pKF292[hupS-egfp] | This work |
| K310 | ΔwhiH::ermE hupS::pKF292[hupS-egfp] | This work |
| K311 | ΔwhiH::ermE ΔhupS::Ωhyg | This work |
| M145 | Prototrophic, SCP1− SCP2− Pgl+ | 29 |
| E. coli strains | ||
| DH5α | Cloning strain | Lab stock |
| ET12567/pUZ8002 | dam-13::Tn9 dcm-6 hsdM; carries RK2 derivative with defective oriT for plasmid mobilization, Kanr | 29 |
| GM2929 | dam-13::Tn9 dcm-6 hsdR2 | M. Marinus |
| Plasmids | ||
| 1C2 | Cosmid carrying hupS (SCO5556) region of the S. coelicolor chromosome | 45 |
| 7A1 | Cosmid carrying hupS (SCO5556) region of the S. coelicolor chromosome | 45 |
| E59 | Cosmid carrying hupA (SCO2950) region of the S. coelicolor chromosome | 45 |
| pCR-BluntII | Cloning vector | Invitrogen |
| pEGFP-1 | Carrying an EGFP gene with human codon usage preferences, Kanr | Clontech |
| pHP45Ωhyg | Source of Ωhyg resistance cassette | 7 |
| pIJ2925 | pUC18-like cloning vector, Ampr | 25 |
| pOJ260 | Mobilizable vector that does not replicate in S. coelicolor, Aprr | 6 |
| pSET152 | Mobilizable vector that integrates at φC31 attB in S. coelicolor, Aprr | 6 |
| pKF286 | hupS EcoRI-PstI fragment from 7A1 into pIJ2925 | This work |
| pKF287 | hupS BglII fragment from pKF286 into pSET152 | This work |
| pKF288 | ΔhupS::Ωhyg insertion in pIJ2925 (Ωhyg inserted between HindIII sites in pKF286) | This work |
| pKF289 | ΔhupS::Ωhyg insertion moved from pKF288 to pOJ260 | This work |
| pKF290 | hupA amplified with KF133/KF134 and cloned in pEGFP-1 to create fusion | This work |
| pKF292 | hupS amplified with KF136/KF137 and cloned in pEGFP-1 to create fusion | This work |
| pKF293 | hupS amplified with KF136/KF138 and cloned in pEGFP-1 to create fusion | This work |
| pKF294 | hupS promoter amplified with KF252/KF253, cloned in pCR-BluntII | This work |
Kan, kanamycin; Amp, ampicillin; Apr, apramycin.
General molecular techniques.
DNA manipulations and cloning were carried out according to standard methods (47). Phusion DNA polymerase (Finnzymes Oy, Finland) was used in PCRs for constructions of plasmids and deletion mutants. Oligonucleotide primers are listed in Table S1 in the supplemental material. Fluorescent DNA sequencing of plasmid constructions made in this work was done using a BigDye terminator version 3.1 cycle sequencing reaction kit and an ABI Prism 3100 DNA sequencer (Applied Biosystems). Transformation of E. coli strains used a slightly modified version of the RuCl protocol (22). Conjugation from E. coli donors into S. coelicolor and polyethylene glycol-mediated transformation of S. coelicolor protoplasts were carried out as described previously (29).
Construction of a ΔhupS::hyg mutant.
For construction of a hupS (SCO5556) deletion strain, an EcoRI-PstI fragment from cosmid 7A1, containing hupS and flanking chromosomal regions, was cloned into pIJ2925 cleaved with the same enzymes to yield plasmid pKF286. HindIII digestion of pKF286 removed 321 bp, 44 bp from the upstream region and 277 bp from the first part of hupS (Fig. 1A), and a hygromycin resistance cassette, cut out using HindIII from plasmid pHP45Ωhyg, was ligated into this site. From this plasmid, named pKF288, BglII was used to excise a fragment containing the disrupted hupS gene and flanking chromosomal regions and ligate it into the BamHI site of pOJ260, which resulted in pKF289. Plasmid pKF289 was introduced into S. coelicolor by conjugation (29), and putative exconjugants in which allelic exchange had occurred through double-crossover recombination events were selected. The allelic exchange was confirmed by PCR and Southern blotting (data not shown), which showed that hupS had been replaced by the ΔhupS::hyg allele. One of the isolated ΔhupS::hyg mutants was named strain K304.
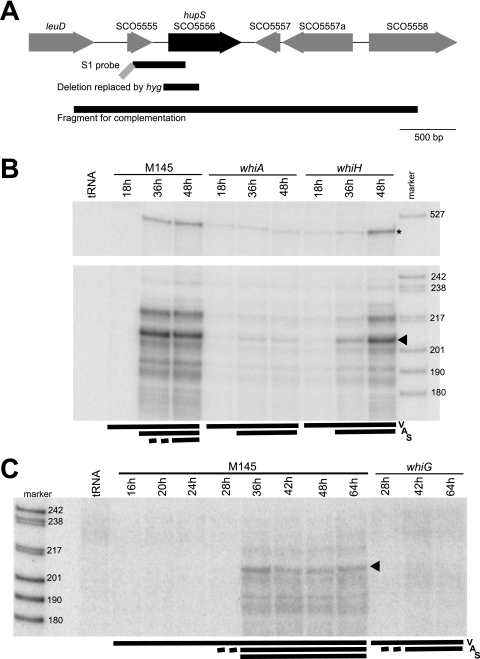
FIG. 1.
(A) Structure of the hupS region of the S. coelicolor chromosome. Locus tags from the S. coelicolor genome sequence are used (5). The extent of the probe used for S1 nuclease assays, the deletion in hupS that was replaced by Ωhyg to generate a knockout mutation, and the fragment that was used in complementation assays are shown below the schematic map as black lines. The gray diagonal line indicates an extension on the S1 probe that contains vector-derived sequence. (B and C) S1 nuclease protection assay of hupS transcripts. Total RNA was extracted from cultures grown on MS agar at the times indicated. The developmental stage at time of harvest is indicated by bars and letters V for vegetative mycelium, A for aerial mycelium covering colony surface, and S for spores being present. An equal weight of RNA was added to each S1 mapping reaction, and a control reaction with an equal weight of yeast tRNA was included in lanes labeled “tRNA.” The lanes labeled “marker” contain a set of DNA molecular weight markers (in base pairs). Panels B and C show different sets of RNA. The RNA preparations represented in panel C are identical to those previously used for mapping ftsZ and smeA promoters (2, 18). Panel B (top) shows protected fragments corresponding to the readthrough of transcription from promoters upstream of the probe (indicated by asterisk). The main protected band is indicated by arrowheads.
For complementation of the ΔhupS::hyg disruption mutant, a 3.1-kb region carrying the intact hupS was excised as a BglII fragment from pKF286 and inserted into pSET152 digested with BamHI, resulting in the plasmid pKF287 (Fig. 1A).
Construction of translational fusions of hupA and hupS to egfp.
The hupA gene was amplified from cosmid E59 using primers KF133 and KF134, which introduced a restriction site for EcoRI upstream of hupA and a BamHI site that removed the stop codon. The hupS gene was similarly amplified using cosmid 1C2 and primers KF136 and KF137, which introduced EcoRI and BamHI sites and removed the translational stop. Digestion with these enzymes and ligation to the corresponding sites in pEGFP-1 resulted in fusion of the reading frames to that of egfp, via codons for amino acid linkers LALDPPVAT for hupA and LQADPPVAT for hupS. The resulting plasmids were named pKF290 and pKF292, respectively. In an alternative construction named pKF293, primers KF136 and KF138 were used to amplify hupS and clone it in pEGFP-1 as described above. The resulting HupS-enhanced green fluorescent protein (EGFP) fusion, in which the last six amino acids of HupS (ATARKK) are replaced by DPPVAT and followed by the EGFP sequence, behaved similarly to the HupS-EGFP fusion produced from pKF292 and gave similar results. Nonmethylated DNA of plasmids pKF290, pKF292, and pKF293 was used to transform S. coelicolor strains into kanamycin-resistant strains, which resulted in integration of the plasmids by homologous recombination at the hupA or hupS locus.
The hupS-egfp fusion was also subcloned on plasmid pSET152 in order to integrate it at the φC31 attachment site in S. coelicolor. However, this resulted in a signal intensity lower than that with integration of pKF292 at the native locus, possibly due to the absence from the pSET152-derived plasmids of putative upstream promoters needed for full hupS expression.
RNA preparation and S1 nuclease protection assays.
Mycelium of S. coelicolor strains grown on cellophane-coated MS agar was harvested three times during development, corresponding to vegetative growth without aerial mycelium (18 h), development of aerial mycelium and initiation of sporulation (36 h), and abundant sporulation leading to gray aerial mycelium (48 h). The investigated whiH and whiA mutants followed a similar development, except that they did not form spores or gray spore pigment. Harvested mycelium was treated with RNAprotect bacteria reagent (Qiagen) to stabilize the RNA. Cell lysis, RNA isolation, and DNase treatment were then carried out using the RNeasy midi kit (Qiagen) according to the total RNA isolation protocol of Mersinias and colleagues (http://www.surrey.ac.uk/SBMS/Fgenomics). In addition, RNA preparations from a previous study were also investigated (18).
S1 nuclease protection assays were carried out essentially as described previously (18). For each reaction, 30 μg of total RNA was hybridized to a probe prepared by PCR. First, a fragment spanning the presumed promoter region upstream of the first start codon was amplified using primers KF252 and KF253 and cloned in a pCR-BluntII-TOPO vector (Invitrogen), resulting in plasmid pKF294. The reverse primer (KF253) was phosphorylated using [γ- 32P]ATP before use in amplification together with a forward primer in the vector sequence to generate a PCR product uniquely labeled on the reverse strand and containing a nonhomologous upstream extension (about 150 nucleotides) to discriminate between full-length protection and probe-probe reannealing products (Fig. 1A). Approximately 30,000 Cerenkov counts min−1 of the labeled probe were used in each hybridization. Digestion with S1 nuclease (Fermentas) was performed for 1 h at 37°C, and digestion products were separated on an 8% denaturing polyacrylamide gel. Molecular weight markers were produced by end labeling of MspI-digested pBR322.
Assays of resistance to heat, detergent, and UV.
Spore suspensions were diluted in water to a suitable concentration and heat treated at 60°C for 5, 10, 20, 30, and 60 min or exposed to 1% or 5% sodium dodecyl sulfate. Serial dilutions were then plated on MS agar plates to determine the number of surviving spores. For assay of UV resistance, serial dilutions of spores were plated on Oxoid nutrient agar and immediately exposed to defined doses of UV irradiation in a Spectrolinker XL-1000 UV cross-linker (Spectronics Corporation) before being incubated at 30°C to determine the number of surviving CFU.
Microscopy.
For observation of GFP signals, vegetative hyphae from liquid cultures were mounted directly on slides coated with 1% agarose in phosphate-buffered saline. Spores or aerial hyphae were sampled by pressing coverslips on the surface of colonies and then placing them on agarose-coated slides. Staining of nucleoids in methanol-fixed spores or aerial hyphae were carried out with 7-amino actinomycin (7-AAD) or 4′,6-diamidino-2-phenylindole (DAPI) as described previously (17). The samples were viewed using a Zeiss Axio Imager.Z1 microscope equipped with X-Cite 120 illumination (EXFO Photonic Solution, Inc.), and images were captured and processed using a 9100-02 electron multiplier-charge-coupled-device camera (Hamamatsu Photonics) or a Photometrics CoolSNAPfx charge-coupled-device camera (Roper Scientific, Inc.) and Volocity 3DM software (Improvision). Stained nucleoids in several images were recognized, and their surfaces measured using the Volocity Quantitation module, after poorly separated or focused nucleoids or unusually large spores with two or more nucleoids worth of DNA had been filtered out. Statistical calculations were done in Microsoft Excel.
Phylogenetic distribution of HupS.
The distribution of genes encoding homologs of HupS in currently available bacterial and archaeal genomes were investigated using BLAST and PSI-BLAST searches at http://www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi.
RESULTS
Developmental regulation of hupS.
A DNA microarray-based investigation of developmentally regulated gene expression during sporulation of S. coelicolor strain M145, which will be reported separately, suggested a transcriptional upregulation of SCO5556, the upregulation being dependent on the developmental regulatory genes whiA and whiH (P. Salerno, J. Larsson, G. Bucca, E. Laing, C. P. Smith, and K. Flärdh, unpublished data). SCO5556 is one of two S. coelicolor genes that encode homologs of the histone-like, nucleoid-associated HU proteins from E. coli and other bacteria. The HU-encoding genes have previously been investigated with the closely related Streptomyces lividans, were designated hup and hup2 (53), and correspond to the S. coelicolor genes SCO2590 and SCO5556, respectively. To better follow conventional genetic nomenclature for bacteria and to indicate that SCO5556 has a specific role during sporulation (see below), here SCO2950 is designated hupA and SCO5556 is designated hupS.
To confirm the result of the DNA microarray study, hupS transcription was monitored using S1 nuclease protection assays. The hupS transcripts were not detected during the vegetative stages of mycelium development but were clearly upregulated at time points when aerial mycelium had developed and spore formation was detectable in the parent strain M145 (Fig. 1B and C). With whiA and whiG mutants, no upregulation of hupS was seen, although faint signals suggested that a very low level of transcription occurred throughout development, at least in the whiA mutant (Fig. 1B and C). In the whiH mutant, the developmental upregulation was delayed compared to the parent strain M145 (Fig. 1 B). These observations show that hupS is developmentally regulated.
Several protected fragments were observed in the S1 nuclease analysis, suggesting that there may be multiple transcriptional starts or mRNA processing sites. Many of the less intense bands may also indicate S1 nuclease-sensitive points in the RNA-DNA hybrids, which complicates the interpretation. The most abundant fragment (Fig. 1B and C) would correspond, as judged from its size, to a hupS transcript with a 5′ end around 45 to 50 bp upstream of the putative start codon of hupS. As shown in Fig. S1 in the supplemental material, a possible promoter motif is present in a suitable position for transcription to start in this region (TTGAtt-17n-TAaCGT, with capital letters matching the proposed consensus for E. coli-like promoters in Streptomyces [8]). This leads us to suggest that this is a putative promoter for hupS. In addition, a larger protected fragment (Fig. 1B) was observed that corresponds to read-through transcription originating over 300 base pairs upstream of hupS. Thus, we cannot rule out the possibility that all the signals are manifestations of transcripts starting upstream of the region covered by the probe. Nevertheless, the results confirm the developmental upregulation of hupS suggested by the DNA microarray analysis and show that this upregulation depends on whiA and whiG and is delayed in a whiH mutant.
The two genes encoding HU proteins in S. coelicolor are differentially expressed during development.
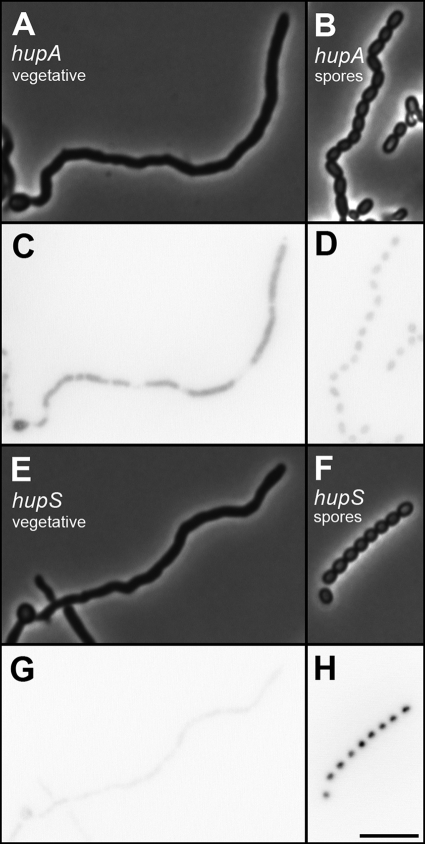
In order to further investigate the temporal and spatial expression of hupS during development, translational fusions of the hupA and hupS coding regions to the gene for EGFP were created. The fusions were integrated by homologous recombination at the corresponding loci in the S. coelicolor strain M145 genome. For both fusion proteins, EGFP signals were observed that appeared to colocalize with nucleoids (Fig. 2). Comparison of the signals in vegetative hyphae and in spores showed that hupA and hupS are differentially regulated. The HupA-EGFP signal was readily detected in vegetative mycelium growing in liquid culture, whereas the signal from HupS-EGFP was very weak (Fig. 2C and G). However, when spores from the surface of developing colonies were investigated, the situation was reversed. The HupS-EGFP signal was significantly upregulated in spores, while the spore signal from HupA-EGFP was relatively weak (Fig. 2D and H). Quantification of the fluorescence signal showed that the average HupA-EGFP intensity value per pixel in spores was about 60% of the value in growing vegetative hyphae (Table 2). On the other hand, the HupS-EGFP intensity for spores was over 10-fold higher than that for vegetative hyphae (indeed, the signal for vegetative hyphae was not significantly different from the value obtained from the inherent autofluorescence of the S. coelicolor hyphae under these conditions).
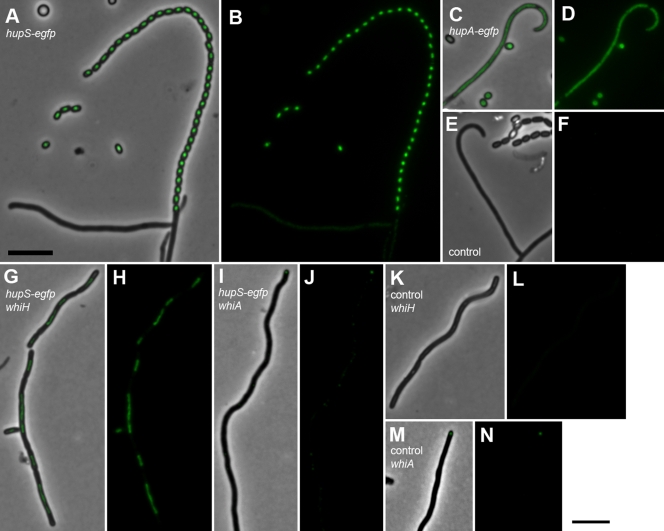
FIG. 2.
Differential regulation of HU-encoding genes hupA and hupS. S. coelicolor strain K306 carrying a hupA-egfp fusion (A to D) and strain K307 carrying a hupS-egfp fusion (E to H) were grown as vegetative mycelium in yeast extract-malt extract liquid medium (A, C, E, and G) or on solid MS agar medium to form spores (B, D, F, and H). Vegetative hyphae and spores were mounted on agarose-coated slides and investigated by phase-contrast and fluorescence microscopy to monitor the signals from the EGFP hybrid proteins. Exposure conditions and image processing were identical for the different samples. Quantification of the fluorescence signals is shown in Table 2. Size bar, 5 μm.
TABLE 2.
Differential expression of hupA-egfp and hupS-egfp in vegetative hyphae and spores
| Strain | Gene fusion | Avg fluorescence intensity (arbitrary unit)a |
Ratiob | |
|---|---|---|---|---|
| Vegetative hyphae | Spores | |||
| K306 | hupA-egfp | 466 ± 168 | 275 ± 74.1 | 0.590 |
| K307 | hupS-egfp | 83.5 ± 40.4 | 948 ± 317 | 11.4 |
Average intensity value per pixel after subtraction of background values from surrounding medium. Measurements were done of 100 randomly selected areas of 0.5 μm2 per strain in images of the type shown in Fig. 2.
Ratio of average fluorescence signal in spores to average signal in vegetative hyphae.
The developmental regulation of hupS-egfp was investigated in more detail in aerial hyphae of wild-type strain M145 and in congenic developmental mutants lacking sporulation regulators whiA and whiH. The upregulation of hupS-egfp was seen with spore chains at different stages of development (Fig. 3A and B and see also Fig. S2 in the supplemental material), but not with subapical stems or with other aerial hyphae in which the signals were barely higher than that of autofluorescence for strain M145 (Fig. 3E and F). As a comparison, hupA-egfp fluorescence was clearly above background autofluorescence in all hyphae in the aerial mycelium preparations, as well as in spores (Fig. 3C and D). The whiH mutant J2408 does not form regular spores, but some aerial hyphae develop into spore-like aerial hyphal fragments in which some sporulation genes are upregulated, as previously documented (16, 17). When introduced into strain J2408, the hupS-egfp fusion showed clear upregulation in such spore-like aerial hyphal fragments, but no clear signal with other hyphae (Fig. 3G and H). On the other hand, the whiA mutant J2401, which makes only long, often tightly coiled, aerial hyphae, showed no detectable signal from hupS-egfp (Fig. 3I and J). We have also investigated a hupS-egfp fusion in whiG mutant J2400 and whiI mutant J2450 without seeing any developmental upregulation (data not shown). Thus, the findings are in agreement with a transcriptional upregulation of hupS in a whiH mutant (Fig. 1B), and this upregulation appears to occur in the aerial hyphal fragments that develop spore-like characteristics. Furthermore, the developmental upregulation of hupS was confirmed to be dependent on whiA, whiG, and whiI.
FIG. 3.
Cell type-specific expression of hupS in sporogenic aerial hyphae of S. coelicolor. Strains K308 (M145 with hupS-egfp; panels A and B), K306 (M145 hupA-egfp; panels C and D), M145 (no egfp; panels E and F), K310 (hupS-egfp in whiH mutant J2408; panels G and H), K309 (hupS-egfp in whiA mutant J2401; panels I and J), J2408 (whiH mutant with no egfp, panels K and L), and J2401 (whiA mutant with no egfp, panels M and N) were grown on MS agar, and aerial mycelium was investigated by fluorescence microscopy. Representative examples of spores or aerial hyphae are shown as a fluorescence image laid over a phase-contrast image (A, C, E, G, I, K, and M) and as only the EGFP fluorescence channel (B, D, F, H, J, L, and N). The weak fluorescent focus at the tip of the hypha shown in panels I and J does likely not correspond to a GFP signal since similar foci were readily detected in the control strain J2401 without any egfp construct (examples shown in panels M and N). Size bar, 5 μm.
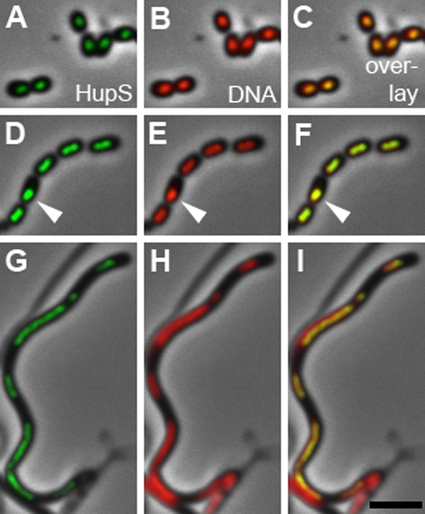
HupS is associated with spore nucleoids.
The subcellular localization of HupS-EGFP signals suggested, as expected for an HU-type protein, that it localized to nucleoids. This was confirmed by staining nucleoids with DAPI in strains expressing hupS-egfp. The EGFP signal clearly colocalized with DAPI-stained DNA (Fig. 4), including in the rare examples of spores in which the nucleoid was aberrantly positioned (Fig. 4 D to F). Furthermore, in the characteristic aerial hyphal fragments of whiH mutants, the nucleoids are partially condensed and irregularly separated, leaving DNA-free gaps between them (17). We found that in these spore-like compartments, all HupS-EGFP signals were similarly restricted to the nucleoid region (Fig. 4 G to I). This confirmed the nucleoid association of HupS in spores.
FIG. 4.
HupS is associated with spore nucleoids. Spores from S. coelicolor strain K307 (A to F) or sporogenic aerial hyphae of whiH mutant K310 (G to I), which both carry a hupS-egfp fusion, were stained with DAPI to visualize nucleoids. The HupS-EGFP signal invariably colocalized with DAPI-stained DNA and also in cases in which the nucleoid was mislocalized (D and F) or irregularly partitioned (G to I). Typical examples of the EGFP signal in green (A, D, and G) or the DAPI signal in red (B, E, and H) projected on phase-contrast images or an overlay of the two fluorescence channels (C, F, and I) are shown. Arrowheads indicate an example of a mislocalized nucleoid to which the HupS-EGFP also localizes. Size bar, 3 μm.
The hupS gene affects spore pigmentation and mounting of heat resistance.
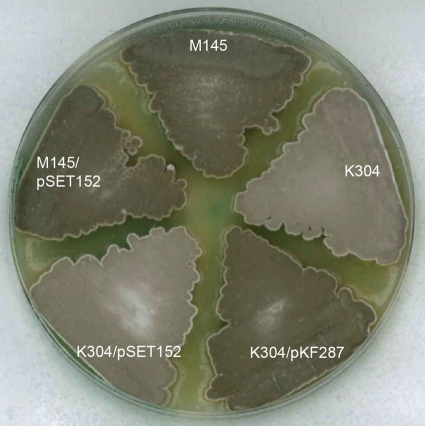
To inactivate hupS, a deletion allele was created in which 44 nucleotides of the upstream region and the 277 first nucleotides of the hupS coding region were replaced by a hygromycin resistance marker (Fig. 1A). This ΔhupS::hyg mutant, strain K304, grew vegetatively and developed aerial mycelium without any detectable differences compared to its parent strain M145. However, the hupS mutant colonies were paler gray than the parent, showing a difference in spore pigmentation (Fig. 5). The mutant spores were normally shaped and of an abundance apparently similar to that of the parent strain (data not shown). Investigation of thin-sectioned mutant spores with transmission electron microscopy showed an apparently normal spore wall in the hupS mutant, with a thickness similar to that of the wild-type spores of strain M145 (data not shown). The pale gray colony phenotype was restored to the normal dark gray of the parent strain when the ΔhupS::hyg mutation was complemented in trans by the hupS region on pKF287 integrated at the φC31 attB site of the S. coelicolor genome (Fig. 5). The complementing plasmid carried a large region upstream of hupS to ensure that the signals for full transcription were included. Since the two genes downstream of hupS are convergently transcribed in relation to hupS, the hupS::hyg mutation should not be polar on the expression on those genes.
FIG. 5.
Plate phenotype and complementation of hupS mutant. The ΔhupS::Ωhyg mutant K304 was grown on MS agar together with its congenic hupS+ parent M145 and derivatives carrying the hupS complementation plasmid pKF287 or the empty vector pSET152 integrated in their genomes.
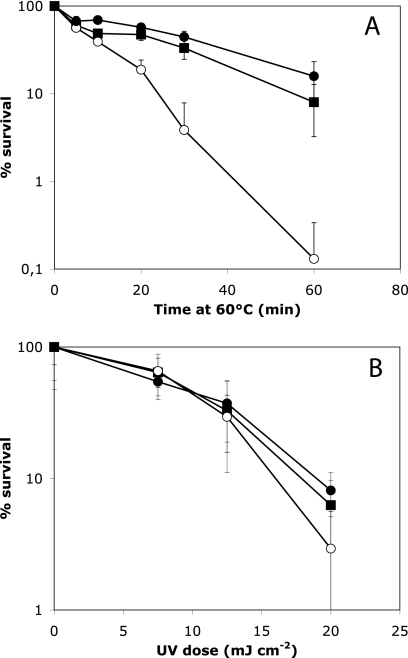
To test whether hupS contributed to the mounting of resistance to stress conditions in spores, we investigated the effects of detergent, heat, and UV irradiation. No significant difference between mutant and parent was detected in the sensitivity of spores to treatment with sodium dodecyl sulfate (data not shown). However, spores of the hupS mutant K304 were clearly less resistant to heat than the spores of the parent strain M145. After incubation at 60°C for 60 min, the survival of spores of the mutant was about 0.1%, which is 100-fold lower than the survival of M145 spores (Fig. 6A). This defect was fully restored by the hupS complementation plasmid pKF287, which resulted in a degree of heat resistance very similar to that of the M145 parent strain (Fig. 6A).
FIG. 6.
Assays of resistance to heat (A) and UV irradiation (B) in spores from the ΔhupS::Ωhyg mutant K304 (open circles), its isogenic hupS+ parent M145 (filled squares), and the mutant strain complemented by hupS in trans in plasmid pKF287 (filled circles). Spores were exposed to 60°C for different times (A) or to different doses of UV irradiation (B), and the numbers of surviving CFU were determined. Mean values and standard deviations are shown, based on data from at least three independent experiments (for panel B, four experiments with three CFU determinations each per UV dose). The hupS mutant spores were clearly impaired in heat resistance but did not show a statistically significant difference in UV resistance compared to the wild-type parent.
One potential role of nucleoid-associated proteins in spores would be to protect DNA from damage or act in DNA repair in order to preserve the genetic information. To test whether HupS may have a role in such processes, we monitored the survival after exposure of spores to UV light. Although a slight increase in the rate of killing of the hupS mutant compared to that of the parent strain or the complemented mutant was apparent, the difference was small and not statistically significant (Fig. 6B).
hupS influences nucleoid structure in spores.
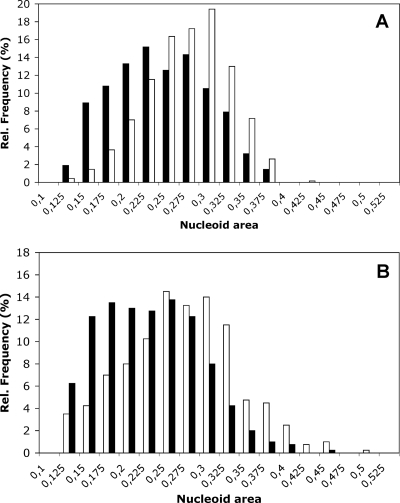
Since other HU-like proteins have been reported to affect nucleoid compaction (41) and since HupS was specifically produced in sporulating cells and was associated with spore nucleoids, we used staining of spore DNA with 7-AAD to find out if it affected nucleoid architecture. The average fluorescently stained nucleoid area of 685 spores of the M145 parent strain was 0.22 μm2, while it was 0.26 μm2 for the hupS mutant K304 (Fig. 7 and Table 3). The average area obtained for 400 spores of the complemented hupS strain K304 with plasmid pKF287 was 0.21 μm2, and for the control strain K304 with pSET152 it was 0.26 μm2. The differences between the hupS+ and hupS mutant strains were in both cases highly significant (P < 0.001) according to Student's t test. Thus, even if measurements of nucleoid areas showed a relatively large degree of variation and the differences between strains were small, the differences were statistically significant. Similar and statistically significant differences between hupS mutant K304 and parent strain M145 or complemented mutant strain K304/pKF287 were reproducibly observed also when nucleoids were stained with DAPI (data not shown).
FIG. 7.
Nucleoid sizes in spores are affected by hupS. Spores from the ΔhupS::Ωhyg mutant K304 (A) (open bars), its congenic hupS+ parent M145 (A) (solid bars), the mutant strain complemented by hupS in trans in plasmid pKF287 (B) (solid bars), and the hupS mutant carrying the empty vector pSET152 (B) (open bars) were stained with 7-AAD, and fluorescence images were collected. Nucleoid areas in normally sized and well separated spores were measured using Volocity 3DM software. Average values and statistics are shown in Table 3. The differences between the hupS+ and hupS mutant strains were in both cases highly significant (P < 0.001) according to Student's t test.
TABLE 3.
Nucleoid areas in wild-type and hupS mutant spores
| Strain | hupS allele(s) | Nucleoid area (μm2)a |
No. of measured spores | |
|---|---|---|---|---|
| Avg | 95% confidence interval | |||
| M145 | hupS+ | 0.22 | 0.223-0.232 | 685 |
| K304 | ΔhupS | 0.26b | 0.258-0.266 | 685 |
| K304/pKF287 | ΔhupS and hupS+ | 0.21 | 0.207-0.219 | 400 |
| K304/pSET152 | ΔhupS | 0.26c | 0.248-0.262 | 400 |
Measured on 7-AAD-stained spores.
Difference from M145 highly significant (P < 0.001) according to Student's t test.
Difference from K304/pKF287 highly significant (P < 0.001) according to Student's t test.
Although average nucleoid areas in spores of the hupS mutant were significantly larger than those in wild-type parental spores, DNA in the mutant spores still appeared to be compacted or condensed to some extent. This was confirmed by investigating a ΔhupS::hyg ΔwhiH::ermE double mutant (strain K311). The spore-like aerial hyphal fragments of whiH mutants are defective in nucleoid segregation but still show condensation of DNA, giving rise to irregularly shaped and sized nucleoids and large DNA-free areas (17). A similar pattern was observed in the hupS whiH double mutant (data not shown), showing that some degree of spore nucleoid compaction occurs also in the absence of hupS.
DISCUSSION
Streptomyces spores do not contain dipicolinic acid or SASPs (5, 14), both of which are main contributors to the remarkable stability of DNA in Bacillus endospores, and the alternative factors that the streptomycetes may use to preserve their DNA and prepare it for long-term survival in spores have not yet been clarified. However, we show here that one of the two HU-type proteins in S. coelicolor, HupS, has a specific role in sporulation, is specifically accumulated in spores, localizes to spore nucleoids, and affects nucleoid size and the heat resistance of spores. Thus, hupS can be added to the list of the relatively few known genes whose gene products have a direct role in the assembly of mature Streptomyces spores (13, 16). This is, as further developed below, likely to be a global role in packaging or protection of the spore DNA. A specific role of hupS related to nucleoids is also indicated by the observation that hupS mutants do not have clear effects on spore wall structure and show normal sensitivity to detergent, which is different from some other previously studied mutants with decreased spore resistance (3, 36, 37, 43). However, at this stage it cannot be excluded that hupS may have an additional or alternative role in controlling expression of specific genes, and this may also contribute to the phenotypes of the hupS mutants. Further investigation of global patterns of gene expression during sporulation will be required to address this. In the meantime, it is clear that HupS accumulates in spores and affects their maturation and nucleoid structure.
Sporulation in S. coelicolor occurs in apical cells of aerial hyphae (16), and the upregulation of hupS appears restricted to such sporogenic cells. In agreement with the compartmentalized expression of hupS in sporogenic cells, spore-like aerial hyphal fragments in whiH mutants also turn on developmental HupS production (Fig. 3G and H). In contrast, hupS depends on the regulatory genes whiA, whiG, and whiI, which all are required for differentiation of recognizable spore-like cells (1, 17). It is unclear whether any of these last regulators have any direct role in controlling the hupS promoter or whether the mutants simply are unable to form the cell type in which hupS is normally upregulated. During division of the sporogenic cell into prespores, partitioning of nucleoids and segregation of a single chromosome into each prespore involves the ParA-ParB-parS system as well as FtsK (24, 52). In addition, segregation is affected by SMC, which also influences nucleoid structure during early stages of sporulation (12, 30). Although it is possible that HupS may also affect these processes during early stages of sporulation, we saw no obvious sign of this. Therefore, we propose that HupS is important mainly at a later stage of sporulation and contributes to DNA packaging and protection in the mature spores. Additional factors will also contribute to the persistence of DNA in the Streptomyces spores, but they still remain to be identified.
HupS has an N-terminal domain of about 90 amino acids that is highly similar to typical HU proteins (see Fig. S3 in the supplemental material), like, for example, E. coli HupA or HupB (38% identity in both cases) and B. subtilis HBsu (40% identity). The 93-amino acid HupA protein in S. coelicolor is also highly homologous to these proteins (e.g., 38% of residues identical to those of E. coli HupA and 42% identity to the N-terminal domain of S. coelicolor HupS). In addition, HupS carries a C-terminal extension of 128 amino acids, which is highly basic and consists of degenerate repetitions of motifs rich in lysines and alanines (see Fig. S3 in the supplemental material). As pointed out previously, such C-terminal repeats are similar to those found in eukaryotic histone H1 (44, 53). HU proteins with such a histone H1-like C-terminal extension appear to be restricted to the Actinobacteria, and we have not found any examples outside of this phylum. A mycobacterial member of this unusual type of HU protein has previously been investigated; Hlp in Mycobacterium smegmatis is present in low abundance in exponentially growing cells (about 120 molecules per cell), but in an interesting parallel to the regulation of hupS in S. coelicolor, Hlp is upregulated during cold shock or anaerobiosis-induced dormancy (31, 38, 39, 44). M. smegmatis Hlp is nucleoid associated and binds DNA in vitro with very high affinity (38, 39). Interestingly, both the N-terminal HU domain and the C-terminal lysine-rich domain bind independently to DNA and both contribute to the high affinity. Hlp could also promote DNA end joining in the presence of DNA ligase and could repress transcription by T7 RNA polymerase (38). The low abundance and the fact that mutants lacking Hlp grew normally and were not affected in survival of cold shock or anaerobiosis show that additional proteins must be involved in organizing the chromatin structure in mycobacteria, e.g., homologs of IHF, H-NS and Dps, and the newly described DNA-bridging protein Lsr2 (10, 31, 49). Hlp was instead suggested to be involved in repair or recombination processes, like nonhomologous end joining, and in regulating gene activity in response to stressful conditions (38).
Our results reveal a strong upregulation of hupS during sporulation, and the clearly detectable HupS-EGFP signal indicates that this is not a low-abundance protein in spores. We show also that HupS has an effect on overall nucleoid structure in spores. Since the positively charged lysine-rich repeats in the C-terminal domains of histone H1 interact with DNA and contribute to the condensation of eukaryotic chromatin (26, 34, 46) and since the corresponding domain in mycobacterial Hlp was found to bind DNA (38), we propose that HupS contributes to chromatin structure and compaction in Streptomyces spores. Intriguingly, it has been proposed that the C-terminal domain of linker histones may derive evolutionarily from bacterial DNA-binding proteins in which the characteristic degenerate alanine- and lysine-rich repeats form proline-kinked α-helical structure that interacts with DNA (11, 26). Presumably, the HupS-like proteins could have arisen from gene fusion between a regular HU-type gene and the gene for such a lysine-rich DNA binding protein. Addition of this extra DNA-binding domain is likely to have changed the interaction of the HU protein with DNA and its effect on chromatin structure.
Since S. coelicolor has two types of HU proteins with different developmental roles, we wanted to clarify the phylogenetic distribution of these kinds of proteins among available genomic sequences from Actinobacteria. Homology searches showed that both types of HU proteins are relatively widespread within the phylum, but that many Actinobacteria have only one of the two types (see Table S2 in the supplemental material). All investigated mycobacterial genomes, as well as representatives of Saccharopolyspora, Nocardia, and Rhodococcus, encode only a long HupS-like HU with a C-terminal extension (see Fig. S3 in the supplemental material). Only a single conventional HU gene (HupA type encoding 90 to 95 amino acids without the histone H1-like C-terminal part) was found in, e.g., Micrococcus, Frankia, Arthrobacter, Bifidobacterium, and Thermobifida. However, eight investigated Streptomyces genomes (including S. avermitilis and S. griseus) encode both a short HU (HupA) and a long one (HupS). In addition, two genes, one for each HU type, were found in Janibacter sp. HTCC22649 and Salinispora tropica (although Salinispora arenicola has only a long HU protein). Finally, as further discussed below, Kineococcus radiotolerans represents an unusual case, with four paralogous genes of the long HupS protein type and a single gene for the short HupA type. Comparison of this distribution to a phylogenetic tree for the Actinobacteria based on 16S rRNA gene sequences does not give a clear picture of when the unusual HupS type of HU proteins may have arisen (51). It is found in Streptomyces and Mycobacterium, but also in relatively distantly related Nocardioides, Salinispora, Saccharopolyspora, and Janibacter. Presumably, it may have appeared early and coexisted with a conventional HU protein, and both were then differentially lost in several lineages. Interestingly, we see no HU homologue at all in the currently available Corynebacterium genomes, suggesting that both genes have been lost in this lineage (unless corynebacteria branched off before a hupS-like gene arose; but its ancestor is highly likely to have carried at least a conventional HU gene, which must have been lost or diverged too far to be recognized). In addition to these scenarios, lateral gene transfer could also have played a role in the phylogenetic distribution of HU-type proteins in Actinobacteria.
The presence of four paralogous hupS-like genes in Kineococcus radiotolerans strain SRS30216 is a possible indicator of a role of HupS-like proteins in protecting DNA. This strain was isolated from a high-level radioactive waste cell and exhibits remarkable resistance to ionizing γ-radiation and to desiccation (42). Desiccation leads to increased frequencies of double-stranded DNA breaks, and desiccation resistance is related to resistance to radioactivity, as indicated by studies of Deinococcus radiodurans (35). Interestingly, although HupS-like proteins were not found outside of the phylum Actinobacteria, the famously radiation-resistant D. radiodurans has an HU protein with lysine-rich repeats in an N-terminal extension, instead of at the C terminus, and this N-terminal part contributes to DNA-binding (20). It remains to be tested whether the HupS-like proteins contribute to radioresistance and desiccation resistance in K. radiotolerans and if they might also do so in S. coelicolor spores or dormant mycobacteria.
Supplementary Material
Acknowledgments
We thank Pelle Wistrand for assistance in constructing some of the EGFP fusions and Nora Ausmees, Stuart Cantlay, and Keith Chater for critically reading the manuscript.
This work was supported by a postdoctoral stipend from Carl Tryggers Foundation to P.S. and by grants from the Swedish Research Council (no. 621-2004-4454 and 621-2007-4767) to K.F. The foundations Crafoordska Stiftelsen, Carl Tesdorpfs Stiftelse, and Maja och Erik Lindqvists Forskningsstiftelse are gratefully acknowledged for support that allowed purchase of the fluorescence microscopy system.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aínsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34:607-619. [DOI] [PubMed] [Google Scholar]
- 2.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 3.Ausmees, N., H. Wahlstedt, S. Bagchi, M. A. Elliot, M. J. Buttner, and K. Flärdh. 2007. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol. Microbiol. 65:1458-1473. [DOI] [PubMed] [Google Scholar]
- 4.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 7.Blondelet-Rouault, M.-H., J. Weiser, A. Lebrihi, P. Branny, and J.-L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the Ω interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 8.Bourn, W. R., and B. Babb. 1995. Computer assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 23:3696-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chater, K. F., and G. Chandra. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651-672. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. M., H. Ren, J. E. Shaw, Y. J. Wang, M. Li, A. S. Leung, V. Tran, N. M. Berbenetz, D. Kocincova, C. M. Yip, J. M. Reyrat, and J. Liu. 2008. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 36:2123-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchill, M. E., and A. A. Travers. 1991. Protein motifs that recognize structural features of DNA. Trends Biochem. Sci. 16:92-97. [DOI] [PubMed] [Google Scholar]
- 12.Dedrick, R. M., H. Wildschutte, and J. R. McCormick. 2009. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 191:320-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliot, M. A., M. J. Buttner, and J. R. Nodwell. 2008. Multicellular development of Streptomyces, p. 419-438. In D. E. Whitworth (ed.), Myxobacteria. Multicellularity and differentiation. ASM Press, Washington, DC.
- 14.Ensign, J. C. 1978. Formation, properties, and germination of actinomycete spores. Annu. Rev. Microbiol. 32:185-219. [DOI] [PubMed] [Google Scholar]
- 15.Flärdh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564-571. [DOI] [PubMed] [Google Scholar]
- 16.Flärdh, K., and M. J. Buttner. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36-49. [DOI] [PubMed] [Google Scholar]
- 17.Flärdh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 18.Flärdh, K., E. Leibovitz, M. J. Buttner, and K. F. Chater. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38:737-749. [DOI] [PubMed] [Google Scholar]
- 19.Frenkiel-Krispin, D., I. Ben-Avraham, J. Englander, E. Shimoni, S. G. Wolf, and A. Minsky. 2004. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 51:395-405. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, S., and A. Grove. 2006. The Deinococcus radiodurans-encoded HU protein has two DNA-binding domains. Biochemistry 45:1723-1733. [DOI] [PubMed] [Google Scholar]
- 21.Guo, F., and S. Adhya. 2007. Spiral structure of Escherichia coli HUαβ provides foundation for DNA supercoiling. Proc. Natl. Acad. Sci. USA 104:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Jakimowicz, D., S. Mouz, J. Zakrzewska-Czerwinska, and K. F. Chater. 2006. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 188:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakimowicz, D., P. Zydek, A. Kois, J. Zakrzewska-Czerwinska, and K. F. Chater. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65:625-641. [DOI] [PubMed] [Google Scholar]
- 25.Janssen, G. R., and M. J. Bibb. 1993. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124:133-134. [DOI] [PubMed] [Google Scholar]
- 26.Kasinsky, H. E., J. D. Lewis, J. B. Dacks, and J. Ausio. 2001. Origin of H1 linker histones. FASEB J. 15:34-42. [DOI] [PubMed] [Google Scholar]
- 27.Kelemen, G. H., P. Brian, K. Flärdh, L. C. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelemen, G. H., G. L. Brown, J. Kormanec, L. Potúcková, K. F. Chater, and M. J. Buttner. 1996. The positions of the sigma factor genes whiG and sigF in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 21:593-603. [DOI] [PubMed] [Google Scholar]
- 29.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 30.Kois, A., M. Swiatek, D. Jakimowicz, and J. Zakrzewska-Czerwinska. 2009. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J. Bacteriol. 191:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, B. H., B. Murugasu-Oei, and T. Dick. 1998. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol. Gen. Genet. 260:475-479. [DOI] [PubMed] [Google Scholar]
- 32.Lee, K. S., D. Bumbaca, J. Kosman, P. Setlow, and M. J. Jedrzejas. 2008. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc. Natl. Acad. Sci. USA 105:2806-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luijsterburg, M. S., M. C. Noom, G. J. Wuite, and R. T. Dame. 2006. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156:262-272. [DOI] [PubMed] [Google Scholar]
- 34.Luijsterburg, M. S., M. F. White, R. van Driel, and R. T. Dame. 2008. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43:393-418. [DOI] [PubMed] [Google Scholar]
- 35.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazza, P., E. E. Noens, K. Schirner, N. Grantcharova, A. M. Mommaas, H. K. Koerten, G. Muth, K. Flärdh, G. P. van Wezel, and W. Wohlleben. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60:838-852. [DOI] [PubMed] [Google Scholar]
- 37.Molle, V., W. J. Palframan, K. C. Findlay, and M. J. Buttner. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Steptomyces coelicolor A3(2). J. Bacteriol. 182:1286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee, A., G. Bhattacharyya, and A. Grove. 2008. The C-terminal domain of HU-related histone-like protein Hlp from Mycobacterium smegmatis mediates DNA end-joining. Biochemistry 47:8744-8753. [DOI] [PubMed] [Google Scholar]
- 39.Mukherjee, A., P. J. DiMario, and A. Grove. 2009. Mycobacterium smegmatis histone-like protein Hlp is nucleoid associated. FEMS Microbiol. Lett. 291:232-240. [DOI] [PubMed] [Google Scholar]
- 40.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paull, T. T., and R. C. Johnson. 1995. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 270:8744-8754. [DOI] [PubMed] [Google Scholar]
- 42.Phillips, R. W., J. Wiegel, C. J. Berry, C. Fliermans, A. D. Peacock, D. C. White, and L. J. Shimkets. 2002. Kineococcus radiotolerans sp. nov., a radiation-resistant, gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:933-938. [DOI] [PubMed] [Google Scholar]
- 43.Potúcková, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 17:37-48. [DOI] [PubMed] [Google Scholar]
- 44.Prabhakar, S., P. S. Annapurna, N. K. Jain, A. B. Dey, J. S. Tyagi, and H. K. Prasad. 1998. Identification of an immunogenic histone-like protein (HLPMt) of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:43-53. [DOI] [PubMed] [Google Scholar]
- 45.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map of the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 46.Roque, A., I. Iloro, I. Ponte, J. L. Arrondo, and P. Suau. 2005. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J. Biol. Chem. 280:32141-32147. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Setlow, P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172-180. [DOI] [PubMed] [Google Scholar]
- 49.Shires, K., and L. Steyn. 2001. The cold-shock stress response in Mycobacterium smegmatis induces the expression of a histone-like protein. Mol. Microbiol. 39:994-1009. [DOI] [PubMed] [Google Scholar]
- 50.Thanbichler, M., and L. Shapiro. 2006. Chromosome organization and segregation in bacteria. J. Struct. Biol. 156:292-303. [DOI] [PubMed] [Google Scholar]
- 51.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L., Y. Yu, X. He, X. Zhou, Z. Deng, K. F. Chater, and M. Tao. 2007. Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2). J. Bacteriol. 189:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama, E., K. Doi, M. Kimura, and S. Ogata. 2001. Disruption of the hup gene encoding a histone-like protein HS1 and detection of HS12 of Streptomyces lividans. Res. Microbiol. 152:717-723. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerman, S. B. 2006. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 156:255-261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.