Abstract
Mycoplasma genitalium is the smallest self-replicating organism and a successful human pathogen associated with a range of genitourinary maladies. As a consequence of its restricted genome size, genes that are highly conserved in other bacteria are absent in M. genitalium. Significantly, genes that encode antioxidants like superoxide dismutase and catalase-peroxidase are lacking. Nevertheless, comparative genomics has revealed that MG_454 of M. genitalium encodes a protein with putative function as an organic hydroperoxide reductase (Ohr). In this study, we found that an M. genitalium transposon mutant that lacks expression of MG_454 was sensitive to killing by t-butyl hydroperoxide and cumene hydroperoxide. To understand whether this sensitivity to hydroperoxides was linked to MG_454, we cloned this gene behind an arabinose-inducible PBAD promoter in plasmid pHERD20T and transformed this construct (pHERDMG454) into a Pseudomonas aeruginosa strain having deletion in its ohr gene (ohr mutant) and showing sensitivity to organic hydroperoxides. The P. aeruginosa ohr mutant harboring pHERDMG454, when induced with arabinose, was able to reverse its sensitivity to organic hydroperoxides, thus supporting the notion that the product of MG_454 resists organic hydroperoxides in M. genitalium. Surprisingly, real-time reverse transcription-PCR showed that expression of MG_454 in M. genitalium was not elevated in response to oxidative stress but was elevated in response to physical stresses, like salt (NaCl) and heat. Although failure of MG_454 to respond to oxidative stress in M. genitalium implies the absence of a known oxidative stress response regulator in the genome of M. genitalium, elevated expression of MG_454 due to physical stress suggests its control by an unidentified regulator.
Aerobic respiration in living organisms leads to the production of superoxide (O2·−) as a by-product of metabolism (12), which subsequently reacts with organic and inorganic molecules to produce oxidative radicals such as hydrogen peroxide (H2O2), hydroxyl radicals (HO·), organic hydroperoxides, and others. These oxidative radicals are often referred as reactive oxygen species (ROS) (36). ROS are highly toxic and cause severe and sometimes irreversible damage to macromolecules like DNA, lipids, and proteins. Therefore, ROS constitute part of the immune defense against invading pathogens, and host phagocytic cells are equipped to specifically produce ROS through special metabolic pathways (11).
Bacteria, regardless of their pathogenic or nonpathogenic nature, have developed strategies to sense and detoxify ROS and to regulate the proteins involved in the detoxification of ROS (18, 28). Such proteins are, in general, enzymes that belong to the oxidoreductase category, which reduces oxide/peroxide groups into corresponding alcohols. Superoxide dismutase (SOD; like Fe-SOD, Mn-SOD, Cu-Zn-SOD), catalase (KatA, KatB), catalase-peroxidase (KatG), alkyl hydroperoxide reductase C (AhpC; a member of the peroxiredoxins [Prx]), and other Prx are common antioxidative enzymes in bacterial species. Expression of these enzymes in response to oxidative stress is controlled at the transcriptional level by specific regulators, such as OxyR (18, 28, 34). For example, OxyR regulates over 30 genes in Escherichia coli, including katG and ahpC (35). Other bacterial regulators associated with control of oxidative stress response proteins are SoxRS (23), PerR (18), Fur (38), and alternate sigma factors (31).
In addition to AhpC, some bacteria possess a distinct enzyme designated organic hydroperoxide reductase (Ohr), which detoxifies organic hydroperoxides. Ohr is a thiol peroxidase, and the gene encoding Ohr was first identified in Xanthomonas campestris by complementing the ahpC mutant strain of Escherichia coli deficient in the regulation of organic peroxide stress (19). Like AhpC and other Prx, Ohr contains two highly conserved cysteine residues that play critical roles in the reduction of peroxides (4). It has been reported that ohr deletion mutants of X. campestris (19), Bacillus subtilis (9), Enterococcus faecalis (29), and Pseudomonas aeruginosa (24) demonstrate hypersusceptibility to hydroperoxide stress compared to their parental strains. Furthermore, ohr is one of the genes isolated from Actinobacillus pleuropneumoniae by in vivo expression technology (32), which is designed to identify genes of pathogenic bacteria that are important for infection but not for in vitro survival. In addition, increasing evidence also indicates that ohr expression in bacteria is upregulated in response to organic hydroperoxide stress (3, 9, 19, 24) and negatively controlled by a regulatory protein called OhrR (3, 14, 26), a hydroperoxide-inducible transcription repressor. Moreover, recent studies have indicated that Ohr proteins are both structurally and functionally similar to the previously reported osmotically inducible protein C (OsmC) of bacteria, and Ohr and OsmC constitute two subfamilies of the Ohr/OsmC superfamily (1, 15, 16).
Mycoplasmas are cell wall-less bacteria of the class Mollicutes and are believed to have originated from the Firmicutes taxon by massive genome reduction to adapt to simple lifestyles (10). The outcome of the reduced genome size in mycoplasmas was the elimination of many highly conserved genes present in other bacterial species. Genes encoding regulatory proteins and oxidative stress responses are typical examples of such selective reduction. Consequently, most mycoplasma genome sequences available in the databases, including those of the human pathogens Mycoplasma genitalium and Mycoplasma pneumoniae, reveal the absence of genes encoding antioxidants like SOD, Kat, and AhpC, although genes encoding the Prx family of proteins are detected in the genomes of mycoplasmas like Mycoplasma penetrans and Mycoplasma pulmonis. Interestingly, however, genomes of M. genitalium (MG_427 and MG_454), M. pneumoniae (MPN_625 and MPN_668), and Mycoplasma gallisepticum (MGA_0252 and MGA_1142) possess two ohr/osmC homologs. Although the putative products of these genes are predicted to have redox function, the products of MG_427, MPN_625, and MGA_0252 and those of MG_454, MPN_668, and MGA_1142 constitute two separate clusters of proteins (13). For example, the products of MG_427, MPN_625, and MGA_0252 exhibit more similarity toward OsmC, whereas the products of MG_454, MPN_668, and MGA_1142 show more similarity to Ohr.
M. genitalium is the smallest self-replicating organism known to date and a successful human pathogen (8). M. genitalium is implicated in genitourinary (36) and respiratory (2) infections, and it is likely that this pathogen encounters oxidative stress during the colonization of mucosal epithelium in both environments. Furthermore, mycoplasmas are known to produce ROS as part of their virulence mechanism (17, 37). Thus, the pathways by which M. genitalium protects itself from oxidative stress are of primary importance in terms of both pathogenesis and survival. Previously, we reported that the oxidative repair enzyme methionine sulfoxide reductase A (MsrA) protects M. genitalium against oxidative stress (6). The present study was undertaken to determine the protective role of M. genitalium Ohr (OhrMg) encoded by MG_454 of M. genitalium.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Wild-type M. genitalium strain G37 (passage 12) (36) was cultured in 100 ml of SP-4 broth in 150-cm2 tissue culture flasks at 37°C for 72 h. The M. genitalium ΔMG_454 and ΔMG_262 strains were grown in SP-4 broth containing tetracycline at 7 μg/ml. SP-4 agar plates (0.87% Noble agar) with or without tetracycline were used as solid media. Escherichia coli and P. aeruginosa parental strains were grown in LB broth or LB agar at 37°C. E. coli strains carrying plasmids were grown in LB broth or LB agar containing ampicillin (100 μg/ml), and P. aeruginosa strains carrying plasmids were grown in LB broth or LB agar containing carbenicillin (300 μg/ml) or tetracycline (10 μg/ml) or both. All cultures and growth assays were conducted under aerobic conditions. The sources for bacteria and plasmids are listed in Table 1.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | lacZΔM15 recA1 | Bethesda Laboratories |
| Mycoplasma genitalium | ||
| G37 | Wild type | 36 |
| ΔMG_454 mutant | ΔMG_454::Tn4001 tet | 10 |
| ΔMG_262 mutant | ΔMG_262::Tn4001 tet | 10 |
| Pseudomonas aeruginosa | ||
| PAO1 | Prototrophic wound isolate | 24 |
| PAO1Δohr::Tc | Tcr; PAO1 harboring a 516-bp deletion of the ohr locus | 24 |
| Plasmids | ||
| pCR2.1 | Apr Kmr; TA cloning vector for PCR fragments | Invitrogen |
| pMG454A | pCR2.1 containing MG_454 coding region with EcoRI ends | This study |
| pHERD20T | E. coli-P. aeruginosa shuttle plasmid containing inducible PBAD promoter | H. Yu, Marshall University |
| pHERDMG454 | pHERD20T containing coding region of MG_454 behind PBAD | This study |
| pUCP22 | Apr; multicopy broad-host-range expression vector | 24 |
| pOHR593 | pUCP22 containing the P. aeruginosa ohr gene including the promoter region | 24 |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
DNA manipulations.
Genomic DNA from M. genitalium strains was isolated using the Easy-DNA Kit (Invitrogen). Plasmids from E. coli were isolated using the QIAprep Spin kit (Qiagen). Sequences for M. genitalium genes were downloaded from NCBI databases. Oligonucleotide primers for the amplification of genes (Table 2) and real-time reverse transcription-PCR (RT-PCR) were synthesized at the DNA core facility at The University of Texas Health Science Center at San Antonio (UTHSCSA). Amplification of DNA fragments was performed by PCR, using standard protocols with M. genitalium genomic DNA as a template. The MG_454 region was amplified with primers MG454EX3 and MG454EX4 and cloned into pCR2.1 to generate plasmid pMG454A. This was digested with EcoRI to release MG_454, which was subsequently cloned into a similarly cut plasmid, pHERD20T (generated by H. Yu, Marshall University), to create plasmid pHERDMG454. This plasmid was used to induce MG_454 expression in P. aeruginosa.
TABLE 2.
Primers used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| MG454EX1 | AGAGGATCCAATATGTTATTTAACATTTTTACTAAA |
| MG454EX2 | TTTGGATCCTTTAAATTTAGTTAAAGCTTAATACC |
| MG454EX3 | CGGCTGAATTCCCATATG TTATTTAACATTTTTACTAAAATA |
| MG454EX4 | TTTGAATTCTTTAGTTAAAGCTTAATACCATTTAAA |
| MG453RTP1 | CACCCCTGAAGGTGATTACAA |
| MG453RTP2 | TTGCAACTCACCACCAACTC |
| MG454RTP1 | TTGCACAAACTGAAACTGGCA |
| MG454RTP2 | TGAGAAAAACAAGTTGCATAAGCAG |
| MG455RTP1 | TTGGTGATCCTACTGGCAGA |
| MG455RTP2 | GCACTAAATGCGTCTGTGCT |
| MG16SRTP1 | AGAGGCGAACGGGTGAGTAA |
| MG16SRTP2 | GGCGCACCCTCATCAAATAA |
Southern hybridization.
Chromosomal DNA from M. genitalium strains cut with BglII was separated onto 1% agarose gels and transferred to Zetaprobe membranes (Bio-Rad) by Southern blotting. After UV treatment, membranes were prehybridized for 4 h in a prehybridization solution containing 50% formamide, 0.12 M Na2HPO4, 0.25 M NaCl, 7% (wt/vol) sodium dodecyl sulfate (SDS), and 1 mM EDTA. Probes for Southern hybridization were labeled with [α-32P]dCTP by a random primer method. A 470-bp PCR fragment containing the MG_454 coding region, obtained by using primers MG454EX1 and MG454EX2 (Table 2) and M. genitalium genomic DNA as a template, and a 2.5-kb HindIII fragment containing the gene for gentamicin resistance, obtained from plasmid pISM2061, served as templates for preparing the probes. Labeled probes were hybridized with membranes for 12 h at 42°C. After hybridization, each membrane was washed for 15 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, 0.5× SSC-0.1% SDS at 42°C, and 0.1× SSC-0.1% SDS at 42°C before exposing to X-ray film for autoradiography.
Peroxide sensitivity assay for M. genitalium.
M. genitalium G37, ΔMG_262, and ΔMG_454 strains were grown in SP-4 medium for 3 to 7 days. Mycoplasma cells were harvested by centrifugation and resuspended in phosphate-buffered saline, and cell density was adjusted to an optical density at 600 nm (OD600) of 0.300. One hundred microliters of diluted cultures were spread in triplicates onto SP-4 agar plates incorporated with different concentrations of hydrogen peroxide (H2O2; 0.15 M to 6.0 M), t-butyl hydroperoxide (t-BHP; 0.05 M to 2.0 M), or cumene hydroperoxide (CHP; 0.032 M to 1.3 M) and incubated at 37°C. Positive growth control plates (without oxidants) received 100 μl of cultures from each strain, and negative growth control plates received 100 μl of phosphate-buffered saline. After 5 days of incubation, growth of M. genitalium strains in the plates was assessed visually by color changes due to phenol red. Those showing colors ranging from yellow to orange, similar to those of positive control plates, were graded as growth positive and not sensitive to oxidants, and those showing red color, similar to that of negative control plates, were graded as growth affected and sensitive to oxidants (see Fig. S1 in the supplemental material).
Peroxide sensitivity assay for P. aeruginosa.
Sensitivity of P. aeruginosa strains to peroxide stress was determined by a disk inhibition assay. P. aeruginosa PAO1, P. aeruginosa Δohr::Tc, and strains harboring plasmids were cultured overnight in LB broth and diluted in LB to an OD600 of 0.300. Two hundred microliters of each culture were mixed with 3 ml of LB soft agar and plated onto LB agar plates containing the appropriate antibiotics. Sterile filter paper disks saturated with 10 μl of t-BHP (0.2 M) and CHP (0.65 M) were placed onto the hardened top agar and incubated at 37°C overnight. The diameters of the zones of growth inhibition were recorded and analyzed statistically.
Quantification of gene expression analysis by real-time RT-PCR.
Total RNA from uninduced M. genitalium cultures and from M. genitalium cultures induced by different stresses was isolated as described previously using Tri reagent (20). RNA was treated with DNase I (Invitrogen) prior to use in real-time RT-PCR assays. cDNA from total RNA was synthesized using Superscript (Invitrogen), and real-time RT-PCR analysis was performed with the ABI Prism 7900 sequence detection system and SYBR green chemistry (Applied Biosystems) as previously described (5, 20). Primers for MG_454 and MG16SrrnA (Table 2) were designed using Primer Express v.2.0. Threshold cycle values (CT) in the exponential phase of the amplification of MG_454 and MG16SrrnA were used to determine transcript levels. MG16SrrnA transcripts were used as the normalizer. Changes in the amounts of transcripts were expressed in n-fold and assessed statistically.
Gene expression analysis by RT-PCR.
RT-PCR was performed to analyze the expression of MG_454 and its adjacent genes (MG_453 and MG_455) in the wild-type and ΔMG_454 mutant strains. Isolation of total RNA and cDNA synthesis was performed as described in the previous paragraph. PCR amplification was performed with cDNA as templates, using a routine protocol with primers MG453RTP1 and MG453RTP2 for MG_453, MG454RTP1 and MG454RTP2 for MG_454, and MG455RTP1 and MG455 RTP2 for MG_455 (Table 2).
Statistical analysis.
Data for disk inhibition assays and real-time RT-PCR were analyzed with one-way analysis of variance using GraphPad Prism 4 (GraphPad Software Inc., San Diego, CA). The Newman-Keuls multiple comparison test (33) was used to analyze statistical differences between groups.
RESULTS AND DISCUSSION
Characterization of the MG_454 mutant.
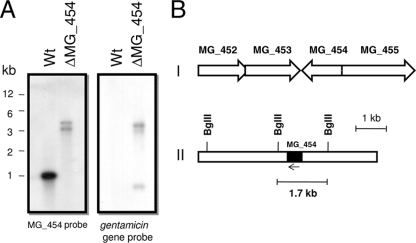
Although M. genitalium possesses two ohr/osmC family genes (MG_427 and MG_454) and their products are predicted to be involved in disulfide reduction (NCBI database), only the product of MG_454 showed significant amino acid sequence identity (23 to 60%) with the Ohr subfamily. A recent phylogenetic analysis (13) also indicated that the product of MG_454 is an ortholog of Ohr. Therefore, we first attempted to determine the role of MG_454 in resistance to oxidative stress. An ΔMG_454 strain was produced (10) as a result of Tn4001 transposon mutagenesis using plasmid pIVT-1 (7). The Tn4001 transposon in pIVT-1 had been modified to possess a tetracycline (tetM) resistance gene in addition to an already existing gentamicin (aacA-aphD) resistance gene. Although the insertion of Tn4001 in MG_454 was previously verified by sequencing using tetM-based primers (10), we performed Southern analysis to further confirm this insertion. Genomic DNA from the M. genitalium wild-type G37 and ΔMG_454 strains was digested with BglII and probed with an MG_454 gene fragment. Hybridization analysis revealed positive signals for MG_454 in DNA from both wild-type and ΔMG_454 strains (Fig. 1A). While the signals in the wild-type strain appeared in the expected size (1,800 bp), signals in the ΔMG_454 strain were higher than those in the wild-type strain. These size shifts confirmed that the Tn4001 insertion occurred within MG_454, and the two bands were due to the presence of additional BglII sites within the inserted sequences (Fig. 1B). To gain additional proof, we probed the wild-type and ΔMG_454 genomic DNA with a 2.5-kb DNA fragment encompassing the gentamicin resistance gene (aacA-aphD) fragment of Tn4001. This probe hybridized only with ΔMG_454 DNA and not with wild-type DNA, further confirming that the ΔMG_454 strain alone contained Tn4001 sequences. Additionally, the signals with this probe in the large-molecular-size region corresponded to the signals obtained with that of the MG_454 probe, thus reinforcing the occurrence of Tn4001 insertion within MG_454. However, the presence of three hybridization signals to the gentamicin resistance gene probe, including one in the small-molecular-size region of less than 1 kb, indicates that there are two internal BglII sites in the vicinity of the gentamicin resistance gene in the plasmid pIVT-1, which is a derivative of pISM2062 (7).
FIG. 1.
(A) Southern hybridization profiles. Wt, wild-type G37 strain; ΔMG_454, ohrMg mutant strain. Sizes of DNA fragments are indicated in kilobases (kb). Chromosomal DNAs were digested with BglII, resolved in 1% agarose gels, Southern transferred to Zetaprobe membranes, and probed with radiolabeled MG_454 or gentamicin gene sequences. (B) Schematics showing the organization of MG_454 in the genome of M. genitalium (I), and locations of BglII restriction sites around MG_454 in the chromosome of M. genitalium (II).
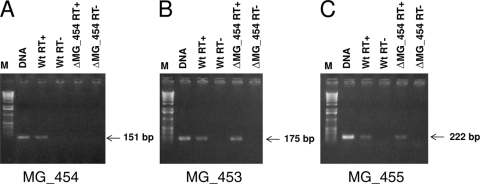
Furthermore, to determine the effect of Tn4001 insertion in MG_454 on the expression of OhrMg in the ΔMG_454 strain, we performed RT-PCR. RT-PCR for MG_454 expression showed no transcript in the ΔMG_454 strain, although it exhibited a significant level of transcripts in the wild-type strain (Fig. 2A). This suggested that the expression of MG_454 is affected in the ΔMG_454 strain. Since transposon insertion sometimes leads to a polar effect on adjacent genes, we also determined the expression of MG_453 and MG_455 by RT-PCR in the ΔMG_454 strain. Results presented in Fig. 2B and C clearly show that expression of MG_453 and MG_455 is unaffected in the ΔMG_454 strain, thus indicating no polar effect on these genes.
FIG. 2.
RT-PCR analysis for M. genitalium genes MG_454 (A), MG_453 (B), and MG_455 (C) from total RNA isolated from M. genitalium wild-type (Wt) and ohrMg mutant (ΔMG_454) strains. PCR products were separated on 1.0% agarose. M, molecular-size marker; DNA, M. genitalium genomic DNA as a template for PCR; RT+, product generated in the presence of reverse transcriptase (Superscript II; Invitrogen) as a template for PCR; RT−, product generated in the absence of reverse transcriptase as a template for PCR. The reaction RT− was done to prove the absence of DNA in total RNAs used for reverse transcriptions.
Sensitivity of the ΔMG_454 strain of M. genitalium to oxidative stress.
The primary role of Ohr is to protect organisms from organic hydroperoxide toxicity. It is very likely, therefore, that organisms lacking Ohr would exhibit a hypersensitive phenotype to hydroperoxides. In order to test this hypothesis, we grew M. genitalium wild-type G37, ΔMG_262, and ΔMG_454 strains in SP-4 agar plates containing different concentrations of H2O2, t-BHP, and CHP. The M. genitalium ΔMG_454 strain failed to grow at intermediate-to-high concentrations of CHP and t-BHP relative to the parental wild-type strain and a control strain (ΔMG_262) bearing the Tn4001 transposon in an unrelated locus, indicating hypersusceptibility to hydroperoxides (Table 3; also see Fig. S1 in the supplemental material). The ΔMG_454 strain also exhibited sensitivity to H2O2 at high concentrations of H2O2 relative to the wild-type strain. These results, which were highly reproducible, implicated OhrMg encoded by MG_454 in the detoxification of oxidants in M. genitalium. Similar results were also reported for ohr mutants of both gram-positive and gram-negative bacteria, like X. campestris (19), P. aeruginosa (24), Bacillus subtilis (9), and Agrobacterium tumefaciens (3). In our previous study (6), we used a disk inhibition assay to demonstrate the sensitivity of msrA deletion mutant of M. genitalium to oxidative stress, and a recent report with M. gallisepticum employed a similar approach to examine mycoplasma sensitivity to oxidants (13). However, in the current study, we found that the growth inhibition assay described in Materials and Methods was much more convenient in determining the effects of oxidants on mycoplasmas, as this is based only on color change (see Fig. S1 in the supplemental material) and does not involve the use of microscopes.
TABLE 3.
Growth of M. genitalium strains in the presence of peroxidesa
| Oxidant concn (M) | Growth of: |
||
|---|---|---|---|
| Wild type | ΔMG_262 mutant | ΔMG_454 mutant | |
| H2O2 | |||
| 1.5 | + | + | + |
| 3.0 | + | + | + |
| 6.0 | + | + | − |
| CHP | |||
| 0.425 | + | + | + |
| 0.650 | + | + | − |
| 1.300 | − | − | − |
| t-BHP | |||
| 0.650 | + | + | + |
| 0.800 | + | + | − |
| 1.000 | + | + | − |
| 2.000 | − | − | − |
+, growth; −, growth affected.
In this study, the concentrations of peroxides and hydroperoxides needed to inhibit the growth of M. genitalium on SP-4 plates appear to be several times greater than those for other bacteria. While M. genitalium require more than 6 M H2O2 to inhibit growth, bacteria like X. campestris (19), P. aeruginosa (24), B. subtilis (9), and Agrobacterium tumefaciens (3) required only 500 mM, 145 mM, 1.6 M, and 1.0 M H2O2, respectively, to inhibit the growth of these bacteria in disk inhibition assays. Similarly, the concentrations of t-BHP (2.0 M) and CHP (1.3 M) required for M. genitalium growth inhibition in SP-4 plates are also a little higher than the concentrations of t-BHP and CHP used for growth inhibition in disk inhibition assays for X. campestris (0.5 M t-BHP, 0.2 M CHP) (19), P. aeruginosa (0.114 M t-BHP, 1.2 M CHP) (24), B. subtilis (0.2 M t-BHP, 0.4 M CHP) (9), and Agrobacterium tumefaciens (1.0 M t-BHP, 0.5 M CHP) (3). The reason for the discrepancy is unclear at present. One possibility could be the nature of the SP-4 medium that we used in this study, which is not only complex but also rich, as it contains 10% fetal calf serum (FBS). It is likely that some metal components in FBS alter the effect of peroxides, thus requiring higher concentrations of peroxides to kill mycoplasmas. Unfortunately, very few growth media exist for mycoplasmas which exclude FBS, and this poses restrictions on testing the sensitivity of mycoplasmas to peroxides in an alternate medium. Aside from this, an alternate explanation may be that mycoplasmas are intrinsically resistant to peroxides. The fact that certain mycoplasmas have the ability to generate and release hydrogen peroxide and superoxide as by-products of normal metabolic activity (17, 37) provides support for this hypothesis. It is possible that the secreted peroxide limits the efficacy of the exogenously added peroxide, leading to the requirement of more peroxide to inhibit the growth. However, this is purely a hypothesis and may not be pertinent to the in vivo situation of mycoplasmas. It is not known at the present whether mycoplasmas that reside inside the host can be killed by peroxide and, if so, the concentration of peroxide that is needed. Nonetheless, it is presumed that OhrMg will at least play a role, since it is functional, to detoxify the hydroperoxides generated by its own metabolism in the in vivo situation. In several intracellular bacteria, which can escape phagocyte-generated peroxide and nitric oxide, antioxidant enzymes like Cu, Zn-SOD, and KatG are still needed for intracellular or in vivo survival (22, 27), and often, they are speculated to play a role in the detoxification of endogenously produced reactive oxygen intermediates.
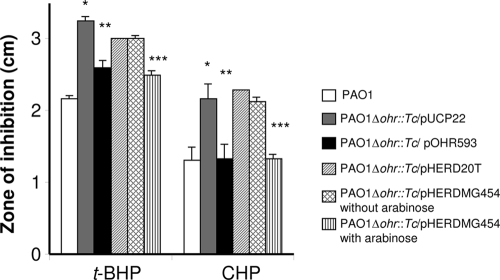
Complementation of the Pseudomonas aeruginosa Δohr::Tc strain with MG_454.
Previous studies have shown that Ohr function in Δohr bacterial strains can be complemented by plasmid-borne ohr (19, 24). Unfortunately, the lack of episomal and integrative plasmids for mycoplasmas restricts such studies. Furthermore, the most direct way in which the ΔMG_454 strain could be complemented would be to clone MG_454 in modified Tn4001 for integration through transposition. However, this approach is very risky because of the promiscuity of Tn4001 and the fact that the ΔMG_454 mutant in the current study was generated using Tn4001 that carries both aacA-aphD and tetM genes, which are the only antibiotic marker genes available for M. genitalium. Hence, complementation of the ΔMG_454 strain was considered unfeasible. In order to offset this situation, we used the P. aeruginosa Δohr::Tc strain as a surrogate host to test complementation by MG_454. The results of the disk inhibition assay (Fig. 3) show that the P. aeruginosa Δohr::Tc strain carrying plasmid pHERDMG454, which has MG_454 fused with the arabinose-inducible PBAD promoter, in the presence of arabinose could reverse the sensitivity of the P. aeruginosa Δohr::Tc strain to CHP and achieve wild-type levels. A more modest but significant correction occurred with t-BHP. These results corroborated those for the P. aeruginosa Δohr::Tc strain harboring plasmid pOHR593 (24) (Fig. 3), which carries the ohr gene of P. aeruginosa, indicating that OhrMg behaves similarly to P. aeruginosa ohr. This finding provides unequivocal evidence that MG_454 encodes an Ohr-like protein. While the Ohr subfamily of proteins from bacteria other than mycoplasmas demonstrates about 40 to 70% identity, Ohr of mycoplasmas show only a limited identity (below 32%) with that of other bacterial genera, such as 25% identity to Ohr of P. aeruginosa. In this context, complementation of the ohr mutant of P. aeruginosa by MG_454, despite low identity, suggests that retention of functional amino acid residues required for the reduction of organic hydroperoxide is more important than overall protein identity. Jenkins et al. (13) have described the presence of functional catalytic cysteine residues and an active site arginine in the Ohr of three mycoplasmas, namely, M. gallisepticum, M. genitalium, and M. pneumoniae; this sequence conservation is similar to Ohr of other species. A glutamate residue that is required to form a salt bridge with arginine to activate the enzyme is also noted to be conserved in these species (15). Thus, the conserved residues may explain the functional complementation of Ohr of P. aeruginosa by OhrMg.
FIG. 3.
Effects of t-BHP and CHP on the growth of P. aeruginosa strains. The disk inhibition method was used to measure the effects of oxidants on the growth of P. aeruginosa strains. Each disk received either 10 μl of 0.2 M t-BHP or 10 μl of 0.65 M CHP. Bars show the diameters of the zone of inhibition in centimeters. PAO1, P. aeruginosa wild-type strain; PAO1Δohr::Tc/pUCP22, P. aeruginosa ohr mutant strain harboring plasmid pUCP22; PAO1Δohr::Tc/pOHR593, P. aeruginosa ohr mutant strain harboring plasmid pOHR593 which carries functional ohr of P. aeruginosa; PAO1Δohr::Tc/pHERD20T, P. aeruginosa ohr mutant strain harboring the plasmid pHERD20T that has the arabinose-inducible PBAD promoter; PAO1Δohr::Tc/pHERDMG454, P. aeruginosa ohr mutant strain harboring the plasmid pHERDMG454 which carries the MG_454 gene behind the PBAD promoter with or without arabinose. * indicates that values are significantly (P < 0.001) higher than the values obtained for PAO1; ** indicates that values are significantly (P < 0.001) lower than the values obtained for PAO1Δohr::Tc/pUCP22; *** indicates that values are significantly (P < 0.001) lower than the values obtained for PAO1Δohr::Tc/pHERD20T and PAO1Δohr::Tc/pHERDMG454 without arabinose.
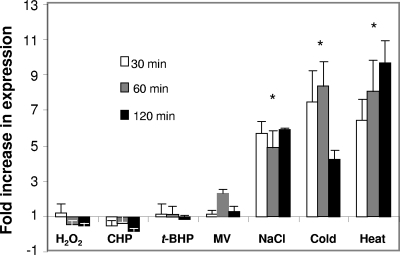
MG_454 is unresponsive to oxidative stress.
Since Ohr encoded by MG_454 provides resistance to oxidants, we explored whether the expression of MG_454 was responsive to exposure to oxidative stress. Real-time RT-PCR results (Fig. 4) showed that transcriptional levels of MG_454 were not affected by H2O2, t-BHP, CHP, and superoxide (methyl viologen) stress. These results corroborate those reported for ohr of M. gallisepticum (13) but differ from that of other bacterial species. For instance, ohr of P. aeruginosa was induced 15- and 5-fold by CHP and t-BHP, respectively (24), and ohr of X. campestris was induced fourfold by t-BHP (19). Interestingly, in B. subtilis, there are two ohr genes, ohrA and ohrB, and only the former is induced by t-BHP and CHP (9). Induction of ohr by CHP has also been reported in A. pleuropneumoniae (32). In many of these species, induction of ohr expression by organic hydroperoxide stress is regulated by a negative regulator, OhrR (3, 9, 26), which functions more or less similarly to that of OxyR, a LysR type of transcriptional regulator that regulates the expression of several bacterial genes, including ahpC, in response to peroxide stress (35). OhrR represses or derepresses the expression of ohr depending upon its redox state. In normal circumstances, OhrR exists in a reduced state, during which it binds to the promoter region of ohr and represses the expression of the latter (3, 9, 26). However, under the conditions of oxidative stress, OhrR is oxidized by oxidants which render it incapable of binding to DNA, thus leading to the derepression of ohr.
FIG. 4.
Real-time RT-PCR determinations of MG_454 expression in response to different stresses. MG_454 transcript levels were determined by real-time RT-PCR as described in Materials and Methods. M. genitalium wild-type strain cultures in log phase (OD600 of 0.600) were subjected to specific stresses (5 mM H2O2, 5 mM CHP, 5 mM t-BHP, 200 mM methyl viologen [MV], 400 mM NaCl, 4°C, and 42°C) for 30 min, 60 min, and 120 min. * indicates that values for all three time points in NaCl, cold, and heat treatments are significantly (P < 0.001) higher than unstressed control values.
In the current study, the absence of induction of MG_454 by oxidants is surprising. Although it appears that this may be due to the lower concentrations of hydroperoxides used in this study, in comparison to the concentrations of hydroperoxides that are required to inhibit the growth of M. genitalium, the concentration of 5 millimolar hydroperoxide/oxidants is proportional to the molar concentrations of these substances required for growth inhibition. This is also similar to other bacteria in which ohr is induced by micromolar concentrations of hydroperoxides and growth is inhibited by millimolar concentrations of hydroperoxides (9, 19, 24). Thus, it appears that absence of induction of the MG_454 gene may be due to lack of an oxidative stress response regulator to regulate MG_454. In line with this, the M. genitalium genome sequence and genome sequences of other mycoplasmas (NCBI database) exhibit no known sequence for genes that are related to oxidative stress response regulators. However, significantly elevated levels of MG_454 expression were observed in response to physical stresses, like NaCl (three- to fivefold), cold (sevenfold), and heat (five- to ninefold). Interestingly, no increase in expression of ohr of M. gallisepticum was detected in response to heat and NaCl stress (13), although a slight induction was observed in response to ethanol stress. In fact, NaCl stress has been reported to downregulate the expression of ohr in M. gallisepticum (13). The B. subtilis ohrB, which is under the control of SigB, an alternate sigma factor, responds to NaCl while ohrA, which is under the regulatory control of OhrR, is unresponsive (9). Since ohr of M. genitalium responds to physical stress, the possibility exists that this gene is controlled by a sigma factor, like that of ohrB of B. subtilis. A similar view has also been suggested for the ohr gene of M. gallisepticum (13). The existence of a gene encoding an RpoE-like alternative sigma factor has been reported in the genomes of M. genitalium, M. pneumoniae, and Ureaplasma urealyticum (21). Whether such a protein plays any role in the regulation of MG_454 remains to be investigated. Furthermore, it appears that MG_454 is not regulated by the heat shock repressor HRCA (heat shock regulation at CIRCE) because the upstream promoter region of this gene lacks the obvious CIRCE (controlling inverted repeats of chaperone expression) element to which HRCA binds for repression of gene expression (20).
Concluding remarks.
Although a previous study has reported that the Ohr homologue of M. gallisepticum possesses organic hydroperoxide reductase activity (13), its role in protecting mycoplasmas against hydroperoxides was not examined. In this study, we provide evidence that OhrMg encoded by MG_454 resists organic hydroperoxide stress in M. genitalium and in a surrogate host, P. aeruginosa. It is very likely that Ohr homologues in the human pathogen M. pneumoniae and poultry pathogen M. gallisepticum have similar functions. This finding is important for the understanding of how mycoplasmas respond to and manage oxidative stress, which has remained elusive. However, it is not clear whether OhrMg is the sole responder to oxidative stress in M. genitalium. Since Ohr is more specific for hydroperoxide reduction, the possibility that M. genitalium and other related mycoplasmas may have other factors to detoxify ROS, like superoxide and hydrogen peroxide, still exists. Alternatively, it is also possible that superoxide and hydrogen peroxide may be converted to hydroperoxides in this species to enable reduction by OhrMg. An additional mechanism may be that methionine residues in proteins act as antioxidants to reduce these compounds, which are later detoxified by enzymes MsrA and MsrB (30). However, key questions that remain to be answered are whether OhrMg plays any role in resisting host-derived ROS or its own ROS while surviving inside the host. If OhrMg is important for host survival, then this protein may serve as a target for drug therapy. The fact that Ohr proteins exist exclusively in bacteria and not in their host makes this hypothesis attractive (25). Finally, the induction of ohrMg (MG_454) by physical stress and not by oxidative stress is interesting. Identification and characterization of the possible regulator for ohrMg may shed additional insights about the role of OhrMg in M. genitalium.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants AI041010 and AI45429.
We are very grateful to John I. Glass, Craig Venter Research Institute, Baltimore, MD, for kindly providing the M. genitalium transposon mutant strains. We also express our gratitude to Michael L. Vasil, University of Colorado Health Sciences Center, Denver, CO, for the P. aeruginosa strains used in this study.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 2.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuchue, T., W. Tanboon, B. Prapagdee, J. M. Dubbs, P. Vattanaviboon, and S. Mongkolsuk. 2006. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J. Bacteriol. 188:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570-11578. [DOI] [PubMed] [Google Scholar]
- 5.Dhandayuthapani, S. 2007. Stress response of genes encoding putative stress signaling molecules of Mycobacterium tuberculosis. Front. Biosci. 12:4676-4681. [DOI] [PubMed] [Google Scholar]
- 6.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 9.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, A., and W. Goebel. 1992. Microbial strategies to prevent oxygen-dependent killing by phagocytes. Free Radic. Res. Commun. 16:137-157. [DOI] [PubMed] [Google Scholar]
- 12.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins, C., R. Samudrala, S. J. Geary, and S. P. Djordjevic. 2008. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J. Bacteriol. 190:2206-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klomsiri, C., W. Panmanee, S. Dharmsthiti, P. Vattanaviboon, and S. Mongkolsuk. 2005. Novel roles of ohrR-ohr in Xanthomonas sensing, metabolism, and physiological adaptive response to lipid hydroperoxide. J. Bacteriol. 187:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesniak, J., W. A. Barton, and D. B. Nikolov. 2002. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 21:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesniak, J., W. A. Barton, and D. B. Nikolov. 2003. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 12:2838-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch, R. E., and B. C. Cole. 1980. Mycoplasma pneumoniae: a prokaryote which consumes oxygen and generates superoxide but which lacks superoxide dismutase. Biochem. Biophys. Res. Commun. 96:98-105. [DOI] [PubMed] [Google Scholar]
- 18.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 19.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musatovova, O., S. Dhandayuthapani, and J. B. Baseman. 2006. Transcriptional heat shock response in the smallest known self-replicating cell, Mycoplasma genitalium. J. Bacteriol. 188:2845-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muto, A., and C. Ushida. 2002. Transcription and translation, p. 323-345. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer/Plenum Publishers, New York, NY.
- 22.Ng, V. H., J. S. Cox, A. O. Sousa, J. D. MacMicking, and J. D. McKinney. 2004. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 52:1291-1302. [DOI] [PubMed] [Google Scholar]
- 23.Nunoshiba, T. 1996. Two-stage gene regulation of the superoxide stress response soxRS system in Escherichia coli. Crit. Rev. Eukaryot. Gene Expr. 6:377-389. [DOI] [PubMed] [Google Scholar]
- 24.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira, M. A., B. G. Guimaraes, J. R. Cussiol, F. J. Medrano, F. C. Gozzo, and L. E. Netto. 2006. Structural insights into enzyme-substrate interaction and characterization of enzymatic intermediates of organic hydroperoxide resistance protein from Xylella fastidiosa. J. Mol. Biol. 359:433-445. [DOI] [PubMed] [Google Scholar]
- 26.Panmanee, W., P. Vattanaviboon, W. Eiamphungporn, W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45:1647-1654. [DOI] [PubMed] [Google Scholar]
- 27.Piddington, D. L., F. C. Fang, T. Laessig, A. M. Cooper, I. M. Orme, and N. A. Buchmeier. 2001. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 29.Rincé, A., J. C. Giard, V. Pichereau, S. Flahaut, and Y. Auffray. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasindran, S. J., S. Saikolappan, and S. Dhandayuthapani. 2007. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2:619-630. [DOI] [PubMed] [Google Scholar]
- 31.Schellhorn, H. E. 1995. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol. Lett. 131:113-119. [DOI] [PubMed] [Google Scholar]
- 32.Shea, R. J., and M. H. Mulks. 2002. ohr, encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect. Immun. 70:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal, R., and F. Rholf. 1981. The principles and practice of statistics in biological research, 2nd ed. W. H. Freeman and Co., New York, NY.
- 34.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 35.Storz, G., and L. A. Tartaglia. 1992. OxyR: a regulator of antioxidant genes. J. Nutr. 122:627-630. [DOI] [PubMed] [Google Scholar]
- 36.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed] [Google Scholar]
- 37.Vilei, E. M., and J. Frey. 2001. Genetic and biochemical characterization of glycerol uptake in Mycoplasma mycoides subsp. mycoides SC: its impact on H2O2 production and virulence. Clin. Diagn. Lab. Immunol. 8:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahrt, T. C., J. Song, J. Siple, and V. Deretic. 2001. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39:1174-1185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.