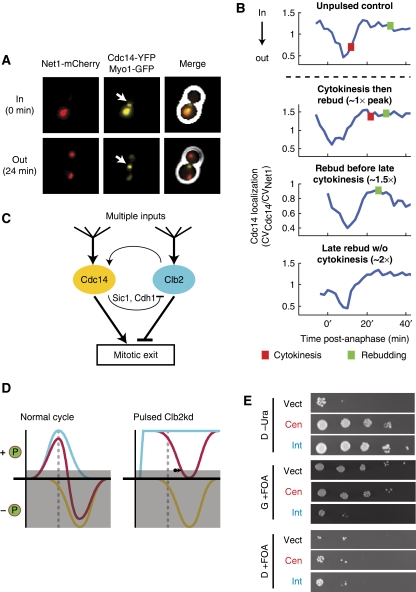
Figure 7.
Mitotic exit may be regulated by kinase/phosphatase balance. (A, B) Cdc14 release from the nucleolus was observed by time-lapse fluorescence microscopy in all cells pulsed with undegradable Clb2. (A) Composite phase contrast, Cdc14–YFP, Net1–mCherry, and Myo1–GFP images of a metaphase-blocked cell (t=0 min) and a cell undergoing release of Cdc14 from the nucleolus (t=24 min). Arrows indicate Myo1–GFP at the bud neck. (B) Cdc14 localization traces at the nucleolus after release from metaphase block for four phenotypes generated by pulsed Clb2kd (YL176-3C). Cdc14 localization was quantified as the ratio of the coefficients of variation (CV=s.d./mean) of pixel intensities for Cdc14–YFP and Net1–mCherry. [Clb2kd] level was estimated by phenotypic correlation with [Clb2kd–YFP] level (Figure 2). (C) Model for co-regulation of mitotic exit by Cdc14 and Clb2–Cdk. (D) Left: normal cells; Clb–Cdk kinase activity (blue) falls after anaphase (dashed line), decreasing phosphorylation of targets. Simultaneous Cdc14 phosphatase release (yellow) further decreases substrate phosphorylation. Mitotic exit occurs when net phosphorylation (red) drops below a threshold (gray box). Right: in the presence of stable Clb2–Cdk activity, Cdc14 can still drive substrate dephosphorylation to allow mitotic exit with a delay (arrow). (E) Additional copies of CDC14 rescue CLB2kd lethality. A diploid strain (ALG876: cdc14/CDC14-YFP CLB2,kd/CLB2 GAL-SIC1(2 × ) ura3/ura3), was transformed with one of three URA3-containing plasmids: vector control (pRS416), centromeric (pRS316-CDC14), or integrating (pRS406-CDC14). Transfomants were grown in galactose and plated in serial dilutions (10-fold) onto glucose (D) or galactose (G) that either lacked uracil (-Ura) or contained 5-FOA. All strains were fully viable on G-Ura (Supplementary Figure 17).