Abstract
Starch defines an insoluble semicrystalline form of storage polysaccharides restricted to Archaeplastida (red and green algae, land plants, and glaucophytes) and some secondary endosymbiosis derivatives of the latter. While green algae and land-plants store starch in plastids by using an ADP-glucose-based pathway related to that of cyanobacteria, red algae, glaucophytes, cryptophytes, dinoflagellates, and apicomplexa parasites store a similar type of polysaccharide named floridean starch in their cytosol or periplast. These organisms are suspected to store their floridean starch from UDP-glucose in a fashion similar to heterotrophic eukaryotes. However, experimental proof of this suspicion has never been produced. Dinoflagellates define an important group of both photoautotrophic and heterotrophic protists. We now report the selection and characterization of a low starch mutant of the heterotrophic dinoflagellate Crypthecodinium cohnii. We show that the sta1-1 mutation of C. cohnii leads to a modification of the UDP-glucose-specific soluble starch synthase activity that correlates with a decrease in starch content and an alteration of amylopectin structure. These experimental results validate the UDP-glucose-based pathway proposed for floridean starch synthesis.
Keywords: alveolate, amylopectin, amylose, starch synthase
Glycogen defines the most widespread form of storage polysaccharides found in archea, bacteria, and eukaryotes. It consists of α-1,4-linked glucan chains hooked together by α-1,6 branches that are distributed symmetrically within small-size hydrosoluble particles. Starch has the same basic composition but contains an heterogeneous mixture of amylopectin and amylose that aggregate into insoluble and semicrystalline granules of unlimited size. Amylopectin, the major fraction of starch, displays a distribution of α-1,6 branches distinctively asymmetrical, thereby allowing this aggregation. By contrast, amylose, which displays very low branching, is dispensable for starch granule formation. While glycogen is widely distributed in eukaryotes, starch is restricted to Archaeplastida (lineages descending from primary endosymbiosis of the plastid) and some secondary endosymbiosis derivatives of the latter. Plastidial starch is found in chloroplasts or amyloplasts of green algae and land plants (the Chloroplastida), while cytosolic starch is found in the cytoplasm of the red algae and glaucophytes (Rhodophyceae and Glaucophyta) and in the cytoplasm or periplast of their secondary endosymbiosis derivatives. The name “floridean starch” has been coined to describe this cytosolic accumulation of storage starch that was described in a particular group of red algae: the Florideophycideae (1).
We have recently proposed that in the common ancestor of the Archaeplastida, starch synthesis occurred in the cytosol. Cytosolic starch metabolism resulted from the merging of the storage polysaccharide pathways of the cyanobacteria-like endosymbiont and of its eukaryotic host, the former using ADP-glucose and the latter UDP-glucose as glucosyl-unit donors (2). Some of the cyanobacterial genes involved in polysaccharide metabolism were transferred to the nucleus and expressed in the cytosol. Both ADP-glucose and UDP-glucose substrates were used in the cytosol for starch synthesis, which thus relied on a mixture of enzymes of cyanobacterial and eukaryotic phylogenies (2). The maintenance of this dual substrate pathway was selected because it allowed for the export of ADP-glucose that was produced in the cyanobiont and its polymerization in the cytosol, thereby achieving the export of photosynthate and establishing the endosymbiotic link (2).
When the three Archaeplastida lineages emerged from this common ancestor, starch was maintained in the cytosol in the glaucophytes and red algae, while it was redirected to plastids that had remained the site of ADP-glucose synthesis in the green algae (the Chloroplastida) (3, 4). The enzymes that were used for elongation of starch in the Chlororoplastida thus relied on those that originally came with the endosymbiont, as these were better adapted to the use of ADP-glucose. On the other hand, we proposed that the glaucophytes and red algae synthesized floridean starch in the cytosol by enzymes of host origin that favored UDP-glucose.
If this proposal is correct, then there should be organisms that produce starch in the cytosol from UDP-glucose using a host-derived glucan synthase. To date there is circumstantial evidence that this is true because an enzyme activity that uses UDP-glucose has been detected (5–9). However, there is no direct correlation between the function of this enzyme and the production of starch. In addition, other reports are suggestive of the presence of an ADP-glucose-based pathway in floridean starch accumulators (1, 10, 11).
This paper provides the missing direct correlation by showing genetic linkage of loss of starch and modification of a UDP-glucose-dependent glucan synthase activity in the model floridean starch accumulator Crypthecodinium cohnii.
Results
Selection of Starch Accumulation Defective Mutants.
Colonies (50,213) that survived UV mutagenesis (at 20% survival) were screened through our iodine staining procedure. A total of 97 mutants were selected, and 32 had a confirmed defect in starch accumulation (Fig. S1). The vast majority of these confirmed mutants (29 out of 32) displayed a yellow phenotype easy to distinguish from the dark blue-black stain of the wild-type. This yellow phenotype correlates with a minimum of 80% decrease in polysaccharide amounts. The three remaining mutants displayed either a red color (strains PP107 and PP406), when sprayed with iodine, or a greenish taint (PP45), suggestive of specific defects, respectively, in amylose and amylopectin synthesis. They accumulated intermediate polysaccharide amounts (between 20 and 90% of wild-type amounts) and will be described elsewhere.
Preliminary Biochemical Characterization and Selection of Strain PP314.
The predominant “yellow” class of mutants was subjected to preliminary biochemical characterization. This consisted in zymogram assays of enzymes of starch metabolism. Four types of activity gels were used (8); these consisted of glycogen-containing polyacrylamide gels incubated with glycosyl-nucleotides (either ADPG or UDPG) or with glucose-1-P to monitor starch synthase and starch phosphorylase, respectively. In addition, starch hydrolases (amylases, glucosidases) and other type of glucan transferases (branching and debranching enzymes, α-1,4 glucanotransferases) can be evidenced in amylopectin or starch containing polyacrylamide gels incubated in buffer without hexose-P or glycosyl nucleotides. In these conditions, the color displayed by the iodine-stained bands will be suggestive of the type of activity revealed (pink for branching enzyme, white for branching enzyme, amylase or glucosidase, blue for debranching enzymes, dark red for α-1,4 glucanotransferases). Only one strain (PP314) repeatedly displayed a strong modification of zymogram pattern. The latter consisted in the replacement of the two fastest migrating bands evidenced in the wild-type reference by a single slower band (Fig. 1A) on glycogen-containing gels incubated with UDP-glucose. This observation was accompanied in crude extracts by a 40% reduction in total assayable UDP-glucose-specific soluble starch synthase activity. We checked all other assayable activities and saw no consistent modifications in other enzyme activities suspected to be involved in starch metabolism (Table S1 and Fig. S2). We therefore embarked on a more detailed genetic and biochemical characterization of the soluble starch synthase defect of strain PP314.
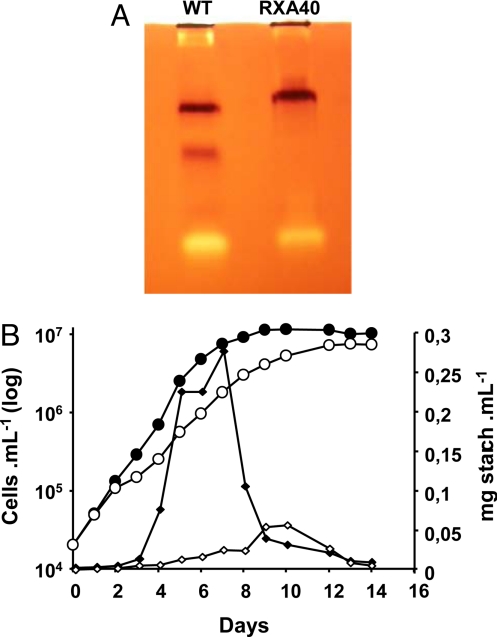
Fig. 1.
Starch deposition is impaired in the PP314 mutant strain. (A) Glycogen-containing zymogram gel incubated with UDP-glucose and stained with iodine. The brown bands correspond to the starch synthase activities detected in the wild-type reference strain (Left) and the PP314 (RXA40) mutant strain (Right). (B) Growth curves and polysaccharide accumulation of wild-type and PP314 mutant strains in liquid rich medium. Growth is displayed as closed black and open white circles, respectively, for the wild-type and mutant strains. The amounts of starch synthesized by C. cohnii wild-type (solid diamonds) and PP314 mutant (open diamonds) strain are displayed and expressed as milligrams of starch per milliliter of culture.
Physiological Characterization of Starch Accumulation, Structural Characterization of the Residual Mutant Starch.
Most yeasts and bacteria accumulate glycogen while approaching stationary phase. Unlike most microorganisms, C. cohnii accumulates storage polysaccharides during early log phase in 1.5% glucose-containing medium, which we have proposed (5) to reflect adaptation of dinoflagellates to a phagotrophic way of life in an otherwise oligotrophic environment (the ocean). They are indeed expected to face sudden burst of nutrients as they ingest prey and are required to rapidly store the bulk of the surplus energy. Inoculation of heterotrophic dinoflagellates in high glucose medium may mimic such bursts. Growth curves and starch accumulation patterns are displayed in Fig. 1B. A 90% decrease in starch amounts relative to the wild-type was scored in liquid-rich medium during log phase. While the wild-type cultures displayed a sharp decrease in starch content as it entered stationary phase, the mutant did not show a comparable decrease and reached a similar very low starch content at the end of stationary phase.
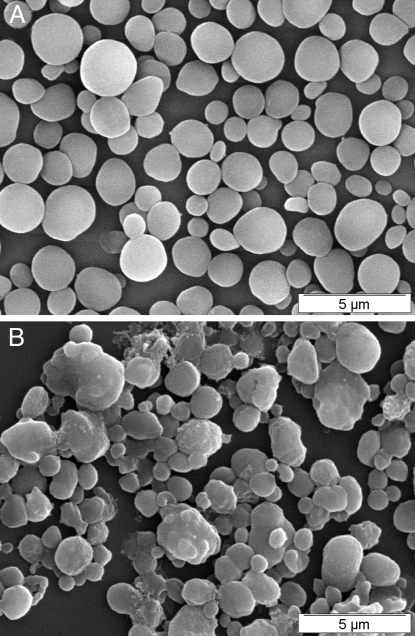
Starch was purified from late log phase wild-type and mutant PP314 cells. The granules were dispersed, and the constituent polysaccharides were separated through gel permeation chromatography. Amylopectin, the high mass component of starch is typically excluded from such columns, while the smaller amylose fraction is clearly separated from the latter and displays a heterodisperse size distribution. The mutant starch was selectively enriched in amylose, which defined 50% of the total starch, while the wild-type accumulated up to 30% of this fraction (Fig. S3). However, on a cell basis, the total amylose content was not modified, taking into account the reduction in starch amounts of the mutant. However, the reverse applies to amylopectin, whose reduction is exacerbated on a total cell basis (70–80% reduction). The color of the iodine polysaccharide interaction is known to depend on the lengths of the glucan chains. The maximal wavelength of the iodine polysaccharide of amylose (λmax) was not significantly modified. However, we did record a significant 10-nm increase in the λmax of amylopectin. We confirmed this modification in amylopectin structure by examining the chain-length (CL) distribution of the wild-type and mutant starch by capillary electrophoresis (CE) of enzymatically debranched chains (Fig. 2). In this technique, the branches are selectively hydrolyzed by an isoamylase. The debranched chains are labeled by fluorescence at the reducing end, thereby yielding the same amount of fluorescence irrespective of their size. The chains are separated by CE, achieving a clear separation for each type of chain differing by as little as one glucose residue up to a degree of polymerization of 70. The CL distribution is thus yielded by detection and quantification of the fluorescence. With this analysis, a decrease in chains ranging between 14 and 30 glucose residues in length was recorded and accompanied by a relative increase in chains ranging between 5 and 13 glucose residues. This modification in amylopectin CL distribution was not restricted to the outer chains of the polysaccharide. Indeed we selectively hydrolyzed such chains through the use of β-amylase and subjected the residual polysaccharide to the same enzymatic debranching and CE analysis of the debranched chains. β-Amylase selectively digests chains from their reducing ends and stops two to three residues from the α-1,6 branch, leaving the core structure with very short external branches. Results displayed in Fig. 2 C and D show that the inner core structure of the polysaccharide was also modified. To assess the consequences of this modification of starch structure on the crystalline packing of the glucan chains, we examined the wide-angle X-ray diffraction (WAXD) pattern of the mutant and compared it to the wild-type. Starches come into two distinct WAXD patterns that correspond to two different elemental packing structures of double helical glucans. A-type diffraction patterns are found in cereal endosperm, plant leaf, and green algae starches. B-type patterns are typical of tuber starches and of starches of high amylose mutants of cereals and algae. While wild-type C. cohnii displayed a pure A-type WAXD pattern (5), the PP314 mutant displayed a mixture of A and B patterns often defined mistakenly as a third C-type diffraction pattern (Fig. S4). Finally we examined starch granule shapes and sizes by scanning electron microscopy on purified granules (Fig. 3). The mutant granule size distribution was more heterodisperse than that of the wild-type. In addition, while wild-type granules appeared ellipsoidal and smooth (Fig. 3A), the mutant starch granule morphology was distinctively altered with a large number of angular, elongated, and sometimes fused granules (Fig. 3B). These alterations are reminiscent of those seen in Chlamydomonas-soluble starch synthase mutants (12, 13).
Fig. 2.
Comparison of the CL distribution profiles for wild-type and mutant amylopectins. After purification on a gel filtration Sepharose CL-2B column, amylopectin was debranched with isoamylase and pullulanase. The resulting glucans were analyzed by CE. The relative proportions for each glucan in the total population are expressed as a percentage of the total number of chains. A and C are, respectively, corresponding to the CL distribution obtained from wild-type amylopectin without and with β-amylase pretreatment. B and D are to the corresponding mutant amylopectin samples. The unbroken lines on B and D correspond to the difference plots obtained by subtracting the mutant CLD to the wild-type CLD.
Fig. 3.
Scanning electron microscopy of native purified starch granules from the wild-type (A) and PP314 mutant (B) strains.
In summary, PP314 displayed a strong reduction in amylopectin content with a significantly modified structure. We can reasonably speculate that the altered starch synthase activity was mostly responsible for the synthesis of the missing amylopectin chains.
Genetic Analysis of the sta1-1 Mutation.
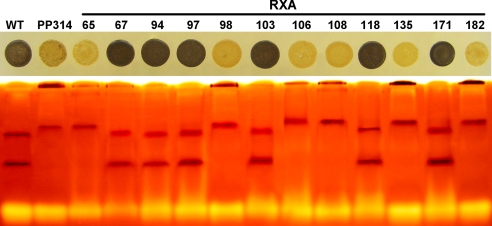
To ensure that the defects in starch structure, polysaccharide amounts, and soluble starch synthase activity resulted from mutation of the same single gene, we embarked in cosegregation analysis of the latter with the genetic defect present in strain PP314. C. cohnii displays a homothallic haplontic life cycle, where the selection and analysis of stable diploid clones is not technically feasible. However, we have recently revisited techniques allowing for gametogenesis, fertilization, and production of meiotic recombinants. In a first step of our analysis, we introduced the canr1-1 marker mutation conveying resistance to 200 μg/mL canavanine by mixing gametes of PP314 with the CAN1 strain. We obtained 3% of our canavanine-resistant clones that simultaneously failed to stain with iodine and therefore appeared yellow, while no such clones were obtained on a total of >1,000 colonies tested when CAN1 was mixed with the ALB strain. This strain displays an albino phenotype, but is otherwise wild-type for starch accumulation. The low starch canavanine-resistant recombinant strain RB1 was then mixed with the ALB strain, and the albino canavanine-resistant recombinant population was analyzed. This enabled us to analyze segregation of the starch accumulation defect in a pure recombinant sample population without contamination by unfused parents. A total of 80 such colonies were stored and subjected to phenotypic characterization. Two discontinuous classes of phenotypes were recorded: 42 Cell patches corresponding to strains accumulating between 80 and 120% of our standard wild-type reference stained black with iodine while 38 strains accumulating from 10–30% of the wild-type amounts stained yellow with iodine. Thirty recombinants were subjected to zymogram analysis (Fig. 4). Among these 30 strains, all low starch recombinants displayed the modified starch synthase zymogram pattern of PP314, while all wild-type recombinants displayed that of our wild-type reference C. cohnii strain. To monitor cosegregation of the starch synthase defect with the alteration in amylopectin structure, we subjected a random sample of the starches purified from three mutant and three wild-type recombinants to the same detailed structural characterization reported above. All three mutants displayed a high amylose content and the same modification in amylopectin CL distribution. These modifications were identical to those evidenced in strain PP314. Taken together, these results prove that PP314 carries a mutation (sta1-1) that behaves as a single Mendelian defect upon crossing. The sta1-1 mutation is responsible for a decrease and modification in UDP-glucose-specific soluble starch synthase activity, which in turn results in a specific decrease in amylopectin synthesis. This decrease is accompanied by significant modification of the structure of both external chains and the internal core of the polysaccharide structure, which in turn is responsible for a modification of the crystalline organization and of the shape and size distribution of the polysaccharide granules.
Fig. 4.
Segregation analysis in the recombinant progeny obtained from the cross between RB1 and ALB1. The colonies of C. cohnii were grown on rich-medium plates and stained with iodine (Upper). The strains displaying a yellow phenotype display a strong (80%) reduction in starch amount. The Lower displays the starch synthases activities detected on glycogen-containing zymograms. The cosegregation between the low-starch yellow phenotype and the modification in starch synthase activities is displayed for 12 recombinant strains harboring both the albinos and the canavanine resistance phenotypes. The two first samples on the Left correspond to the wild-type and the original PP314 mutant strains, respectively.
Biochemical Characterization of the Mutant Soluble Starch Synthase Activities.
To further confirm that the mutant strain contained modified soluble starch synthase activities, we tried to separate the three major activities visualized in crude extract zymograms following the procedure established previously for wild-type strains (5).
The first step took advantage of the difference in affinities for α-glucans observed for each starch synthase isoform on zymograms, with the slowly migrating form having more affinity. It consisted of affinity chromatography on an amylose column. In the wild-type, the slowly migrating starch synthase was retained on this resin, while the fast-migrating form was found back in the low-affinity fraction of the protein mixture allowing further characterizations of the two semipurified fractions (5). The same procedure using a mutant strain crude extract revealed once again a modification of the starch synthase activities in PP314. We were able to detect two different starch synthase activities retained on the amylose column, the slowly migrating form as it was assessed in a wild-type but also a faster migrating isoform (Fig. S5), which however migrated slower than the two wild-type low affinity isoforms. However, a part of this faster-migrating isoform was still found in the unretained fraction, a majority of this activity was coeluting with the slowly migrating isoform. Several chromatographic procedures were tried to separate the two major starch synthase activities (the high and low affinity activities) in the mutant background (anion exchange HiTrapQ column, Gel permeation S300 column), but we always observed coelution of both activities. These results are consistent both with either the selective loss of a particular enzyme or enzyme subunit in PP314 or with the posttranslational modification of starch synthase activities.
Discussion
That heterotrophic eukaryotes polymerize glycogen from UDP-glucose was proved beyond doubt by mutant selection and biochemical analysis in yeasts, fungi, and animals. Similarly, the isolation of starchless or glycogenless mutants of plants, green algae, and bacteria prove that the latter use ADP-glucose only to polymerize their storage polysaccharides. However, the status of floridean starch synthesis was unclear, and experimental reports supporting either an ADP-glucose- or an UDP-glucose-based pathway have appeared (1). To bring functional evidence with respect to this and other issues concerning floridean starch synthesis, we engaged in the selection of mutants defective for the synthesis of such polymers. In our opinion, the only organism among floridean starch accumulators that seemed amenable to such a functional approach consisted of the unicellular heterotrophic dinoflagellate C. cohnii (5). Indeed, >30 years ago, two research groups published the selection of a number of adenine, guanine, or cytosine auxotrophs and of mutants defective for either cell motility or carotenoid biosynthesis in this species (14–17). None of these mutants were sufficiently characterized to uncover a specific biochemical defect, but they were instrumental in setting up conditions for gametogenesis, crossing, and recombinant selection in this model homothallic dinoflagellate species. Nevertheless, these pioneering studies were not pursued, and the original mutants were lost. Moreover in our hands, the different gametogenesis techniques detailed by these two groups failed to trigger the sexual cycle of our reference strain. We have recently reinvestigated this issue and set out to select for mutants defective for floridean starch metabolism in C. cohnii (5). We now describe a low starch mutant of this dinoflagellate species that displays reduced and altered UDP-glucose-specific soluble starch synthase activity. This establishes an UDP-glucose-based pathway for floridean starch synthesis and describes a dinoflagellate mutant with an identified biochemical defect. The existence of such a pathway is consistent with the idea that in the ancestor of the Archaeplastida, both ADP-glucose and UDP-glucose pathways were indeed operating to generate cytosolic starch. This dual pathway was essential in establishing the connection between the eukaryotic host and its endosymbiont. Bioinformatic mining of genomic resources identifies candidate genes for enzymes of glucan elongation in some floridean starch accumulators. These genes are related to sequences found in many diverse nonphotosynthetic eukaryotes where they are thought to be used for glycogen metabolism. According to the CAZy classification (18), the candidate genes encode a GT5-type of glycosyl transferase, which is quite distinct from the intensively studied GT3-type of glycogen synthase known to polymerize glycogen from UDP-glucose in animals and fungi. Both the function of the putative eukaryotic GT5 glycogen synthases and their substrate preferences remain to be ascertained.
The primitive status of genomics and the typical very large size dinoflagellate genome of C. cohnii prevented us from identifying candidate soluble starch synthase genes. Despite the finding of GBSSI-like sequences, we were unable in addition to locate GT5 or GT3 soluble starch synthase-like sequences in Crypthecodinium or other dinoflagellate EST resources. Anyhow, it is quite possible that STA1 does not code the altered starch synthase activity. Indeed in yeast, low glycogen mutants were selected through an analogous mutant screen by forward genetics. The vast majority of mutants selected encoded components of the cell signal transduction machinery that included protein kinases and phosphatases and not the structural genes of enzymes of the glycogen pathway (19). Comparable elements had been known for years to regulate mammalian glycogen metabolism. The altered mobility and kinetics of the modified starch synthase could be interpreted along similar lines.
Materials and Methods
Generation of a Mutant Bank of C. cohnii Defective for Starch Metabolism and Iodine Screening.
The algal C. cohnii strain ATCC40750 was grown and maintained in the dark at 27 °C in liquid and solid rich medium, respectively (5). The UV mutagenesis was performed using a Transilluminator UV LightBox (UV Products) at 302 nm. Colonies were selected through spraying with iodine vapors as detailed in ref. 5. The resulting iodine staining depends on both the amount and composition of the storage glucans. The iodine-polysaccharide complex develops a color depending on the constituent CLs giving a weak brownish color for glycogen and a purple to green stain for, respectively, amylopectin and amylose (20). This procedure allows the identification of low starch amount accumulating strains (yellow), of strains containing starch enriched in amylose (greenish), or accumulating amylopectin only (purple-red).
Cell Crossing.
Crosses were obtained by simply mixing the same amount of the two parental strains that had just reached stationary phase in MLH medium (5, 21) at 15 °C.
Starch Synthase Zymograms, Enzymatic Assays, and Partial Purification Procedures.
The procedure used to detect starch synthase activities on rabbit liver glycogen containing polyacrylamide gels or to assay the enzymatic activity from algal crude extracts has been already detailed in ref. 5. Briefly, the proteins from a crude extract were separated by electrophoresis in native conditions at 4 °C, and the gels were incubated in the presence of UDP-glucose. The staining of the polyacrylamide gels with iodine allowed the detection of dark bands corresponding to the elongation of the outer chains of the embedded glycogen by starch synthase activities. The starch synthase activities in crude extracts were monitored by incubating 20 to 100 μg proteins in the presence of radiolabeled UDP-glucose as described in ref. 7. The partial purification of starch synthase activities from both wild-type and mutant crude extracts were carried out from 2-L algal cultures in midlog phase. The crude extracts were subsequently loaded onto an amylose affinity followed by anion exchange columns using the settings described in ref. 5. The presence of the different starch synthase activities in the semipurified fractions obtained was assessed by zymogram analyses.
Starch Amount Assay and CL Distribution Analysis.
Crypthecodinium cultures (1 L) were inoculated at 25,000 cells/mL with midlog phase liquid precultures of known cell density. The starch determination was performed on 50-mL samples of these cultures following the protocol described elsewhere (5). The amount of starch accumulated was assessed >12 days of growth. The starch amount was measured with respect to the culture volume. This value was also adjusted to cell numbers. Cell counts were determined using a Coulter particle counter. Separation of amylose and amylopectin was performed using 15 mg purified starches onto a CL-2B gel permeation column as described in ref. 7. The GPC-purified amylopectins were enzymatically debranched with 10 units Pseudomonas amylodermosa isoamylase (Hayashibara Biochemical Laboratory) at 42 °C for 12 h. The glucose chains obtained were labeled with 8-amino-1,3,6-pyrenetrisulfonic acid, and the CL distribution determined by CE (22). For the determination of the inner core CL distribution, the same procedure was followed, except that the amylopectins were first digested with 17 units β-amylase from sweet potato (Sigma) in 55 mM sodium acetate, pH 3.5, for 4 h at 30 °C before debranching by isoamylase.
Scanning Electron Microscopy.
Dilute starch granule suspensions were allowed to dry on freshly cleaved, glow discharged mica discs glued on copper stubs. The samples were coated with Au/Pd and observed in secondary imaging electron mode with a JEOL JSM-6100 microscope operating at 15 kV.
Wide-Angle X-Ray Diffraction.
An aliquot of starch granules was centrifuged and the resulting slurry was poured into a 0.7-mm-wide glass capillary. The capillary was sealed, placed in vacuum, and X-rayed with a Ni-filtered CuKa radiation (λ = 1.542 Å), using a Philips PW3830 generator operating at 30 kV and 20 mA. WAXD patterns were recorded on Fujifilm imaging plates, read with a Fujifilm BAS-1800II bioimaging analyzer. Diffraction profiles were calculated by radially integrating the intensity in the two-dimensional patterns.
Supplementary Material
Acknowledgments.
This research was funded by the Agence Nationale de la Recherche grant “GenoFonctDinoFI,” French Ministry of Education, Centre National de la Recherche Scientifique, Région Nord Pas de Calais, and European Union (support to S.B. and J.L.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907424106/DCSupplemental.
References
- 1.Viola R, Nyvall P, Pedersen M. The unique features of starch metabolism in red algae. Proc R Soc Lond B Biol. 2001;268:1417–1422. doi: 10.1098/rspb.2001.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deschamps P, et al. Metabolic symbiosis and the birth of the Plant Kingdom. Mol Biol Evol. 2008;25:536–548. doi: 10.1093/molbev/msm280. [DOI] [PubMed] [Google Scholar]
- 3.Deschamps P, Moreau H, Worden AZ, Dauvillée D, Ball SG. Early gene duplication within Chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts. Genetics. 2008;178:2373–2387. doi: 10.1534/genetics.108.087205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deschamps P, Haferkamp I, d'Hulst C, Neuhaus E, Ball S. The relocation of starch metabolism to chloroplasts: When, why and how. Trends Plants Sci. 2008;13:1802–1816. doi: 10.1016/j.tplants.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Deschamps P, et al. The heterotrophic dinoflagellate Crypthecodinium cohnii defines a model genetic system to investigate cytoplasmic starch synthesis. Eukaryot Cell. 2008;7:247–257. doi: 10.1128/EC.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppin A, et al. Evolution of plant-like crystalline storage polysaccharide in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J Mol Evol. 2005;60:257–267. doi: 10.1007/s00239-004-0185-6. [DOI] [PubMed] [Google Scholar]
- 7.Deschamps P, et al. The nature of the periplastidial pathway of amylose synthesis in the cryptophyte Guillardia theta. Eukaryot Cell. 2006;5:954–963. doi: 10.1128/EC.00380-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plancke C, et al. Pathway of cytosolic starch synthesis in the model glaucophyte Cyanophora paradoxa Eukaryot. Cell. 2008;7:247–257. doi: 10.1128/EC.00373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyvall P, Pelloux J, Davies HV, Pedersen M, Viola R. Purification and characterization of a novel starch synthase selective for uridine 5′-diphosphate glucose from the red alga Gracilaria tenuistipitata. Planta. 2001;209:143–152. doi: 10.1007/s004250050616. [DOI] [PubMed] [Google Scholar]
- 10.Nagashima H, Nakamura S, Nisizawa K, Hori T. Enzymic synthesis of floridean starch in a red alga, Serraticardia maxima. Plant Cell Physiol. 1971;12:243–253. [Google Scholar]
- 11.Sesma J, Iglesias AA. Synthesis of floridean starch in the red alga Gracilaria gracilis occurs via ADP-glucose. In: Garab G, editor. Photosynthesis: Mechanisms and Effects. Vol 5. Dordrecht, The Netherlands: Kluwer Academic; 1998. pp. 3537–3540. [Google Scholar]
- 12.Buléon A, et al. Starches from A to C. Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 1997;115:949–957. doi: 10.1104/pp.115.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buléon A, Colonna P, Planchot V, Ball S. Starch granules: Structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 14.Beam CA, Himes M. Evidence for sexual fusion and recombination in the dinoflagellate Crypthecodinium (Gyrodinium) cohnii. Nature. 1974;250:435–436. doi: 10.1038/250435a0. [DOI] [PubMed] [Google Scholar]
- 15.Beam CA, Himes M, Himelfarb J, Link C, Shaw K. Genetic evidence of unusual meiosis in the dinoflagellate Crypthecodinium cohnii. Genetics. 1977;87:19–32. doi: 10.1093/genetics/87.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuttle RC, Loeblich AR. Genetic recombination in the dinoflagellate Crypthecodinium cohnii. Science. 1974;185:1061–1062. doi: 10.1126/science.185.4156.1061. [DOI] [PubMed] [Google Scholar]
- 17.Tuttle RC, Loeblich AR. N-methyl-N′-nitro-N-nitrosoguanidine and UV induced mutants of the dinoflagellate Crypthecodinium cohnii. J Protozool. 1977;24:313–316. doi: 10.1111/j.1550-7408.1977.tb00985.x. [DOI] [PubMed] [Google Scholar]
- 18.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannon JF, Pringle JR, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banks W, Greenwood C, Khan K. The interaction of linear amylose oligomers with iodine. Carbohydr Res. 1971;17:25–33. [Google Scholar]
- 21.Tuttle R, Loeblich AR. An optimal growth medium for the dinoflagellate Crypthecodinium cohnii. Phycologia. 1975;14:1–8. [Google Scholar]
- 22.Morell MK, Samuel MS, O'Shea MG. Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis. 1998;19:2603–2611. doi: 10.1002/elps.1150191507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.