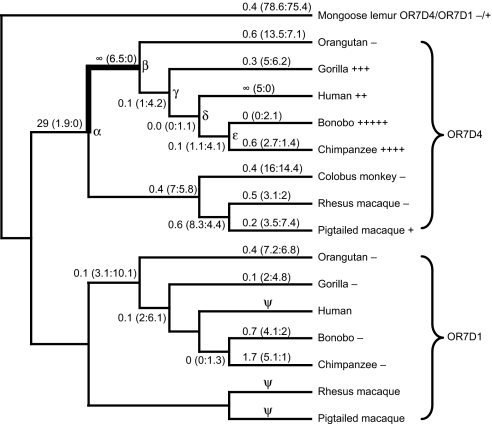
Fig. 2.
OR7D4 shows a signature for positive selection in the primate lineage. The ω values and actual numbers of nonsynonymous and synonymous changes (R:S in parentheses) along each branch are calculated by the free-ratio model of PAML using the OR7D4+OR7D1 dataset and shown on a cladogram of accepted primate phylogeny (36). Bolded line indicates accelerated evolution as computed by likelihood ratio tests shown in Table S4. Hypothetical OR7D4 ancestors, including catarrhine (α), Great Ape (β), hominine (γ), human, bonobo, and chimpanzee (δ), and bonobo and chimpanzee (ε), are indicated on the corresponding nodes on the cladogram. The functional level of the OR7D4 and OR7D1 orthologs for each species as measured in the luciferase assay (see Fig. 1A and Fig. S3A) is indicated beside the species name. The number of plus signs indicates the relative level of functional activity, and the minus signs indicate that the ortholog is nonfunctional in response to androstenone or androstadienone. Ψ denotes pseudogene lineages that are excluded from the analysis.