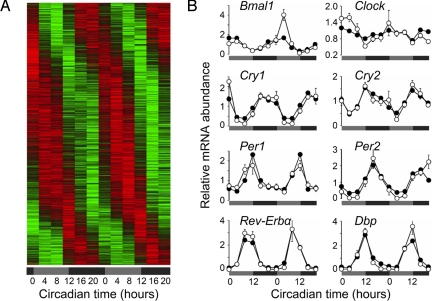
Fig. 4.
Eight percent of all transcripts in macrophages are expressed with a circadian rhythm. (A) Phase-sorted heat map of genes transcribed in a circadian manner in peritoneal macrophages. Cells harvested via peritoneal lavage from four C57BL/6 mice every 4 h were magnetically purified for CD11b surface expression (see Fig. S5). Three individual RNA samples of each time were pooled and subjected to global gene transcription measurement by using Affymetrix chips. The analysis on circadian rhythmicity was done with CircWaveBatch. Lfdrs were determined and cutoff value was arbitrarily set to 0.1 as a measure for rhythmic versus nonrhythmic transcripts (see Fig. S7). Genes expressed in a circadian manner were plotted phase-sorted in a heat-map style (colors indicate min–max normalized relative expression: green, minimum expression; red, maximum expression). (B) Canonical clock gene expression in peritoneal macrophages. Individual datasets from A were plotted (filled circles) together with data obtained by a quantitative RT-PCR assay of the same samples (open circles, means ± SEM, n = 4, except for times CT 24 and 28, n = 3). Statistical analysis for qPCR data and chip data were performed with CircWave and CircWaveBatch, respectively (microarray: P ≤ 0.0001: Bmal1, Cry1, Per2; P ≤ 0.01: Rev-Erbα, Dbp, Cry2; P ≤ 0.05: Per1 and Clock; qPCR: P ≤ 0.0001: Bmal1, Cry1, Per1/2, Clock, Rev-Erbα, Dbp; P ≤ 0.001: Cry2).