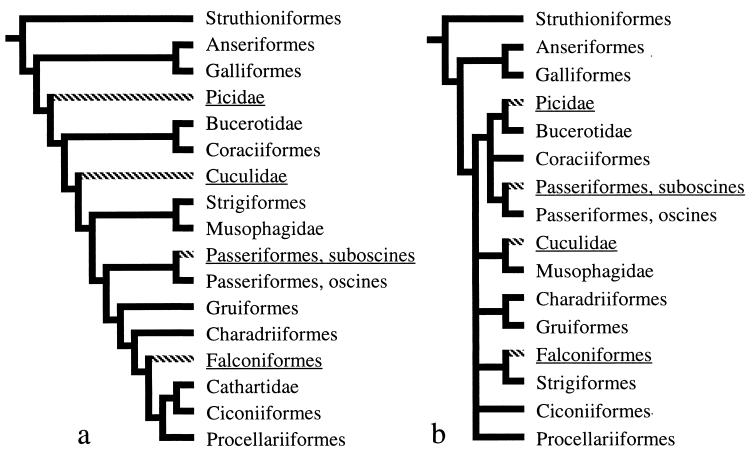
Figure 2.
Most parsimonious distribution of two different mitochondrial gene orders on two avian phylogenetic hypotheses. Gene order previously known for birds (11) (Fig. 1b) is shown with solid branches, and the novel gene order (Fig. 1c) is shown with cross-hatching on branches. Phylogenetic hypotheses follow published analyses by Sibley and Ahlquist based on DNA–DNA hybridization analyses (a) (20) and Cracraft based on morphological characters (b) (21). We consider the published analyses described in a and b to be more comprehensive than analysis of the limited (in taxa and characters) primary sequence data associated with sequencing of gene junctions for this study. Nonetheless, we conducted a series of phylogenetic analyses by using parsimony for the 32 species in Table 1 sequenced for fragment 1, 2, or 4, sharing about 400 bp spanning the 3′ end of Cytb and tRNAT. All analyses, using both equal and various unequal weights for transversion substitutions versus transition substitutions, yielded most-parsimonious trees in which taxa with the novel gene order (Fig. 1c) were nonmonophyletic and scattered throughout the tree. We also conducted analyses with more than 1,900 bp of mitochondrial sequence (from the mt 12S rDNA, Cytb, ND3, and tRNAT genes) for 15 of our Table 1 taxa, including representatives of three of the orders in which we found the novel gene order, using both equal and unequal weighting, and these also support nonmonophyly of taxa with the novel mitochondrial gene arrangement.