Figure 1.
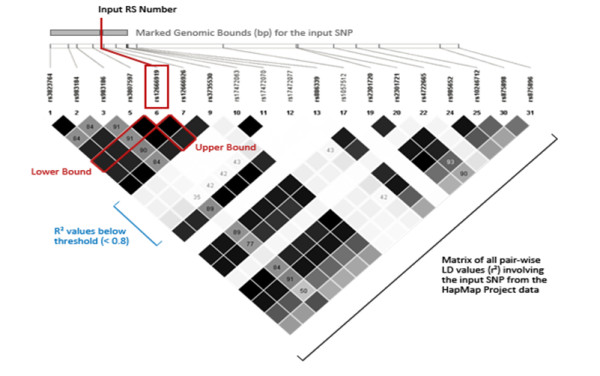
Overview of the LD-Spline Algorithm. A matrix of all HapMap-based pair-wise LD values (D' or r2) is retrieved from the database. Using this matrix, the lower bound is incrementally extended to downstream SNPs while the pair-wise LD value between the downstream SNP and the input SNP is greater than the user-defined threshold (in this case r2 > 0.8). The process is repeated for the upper bound to define the marked genomic bounds for the input SNP.
