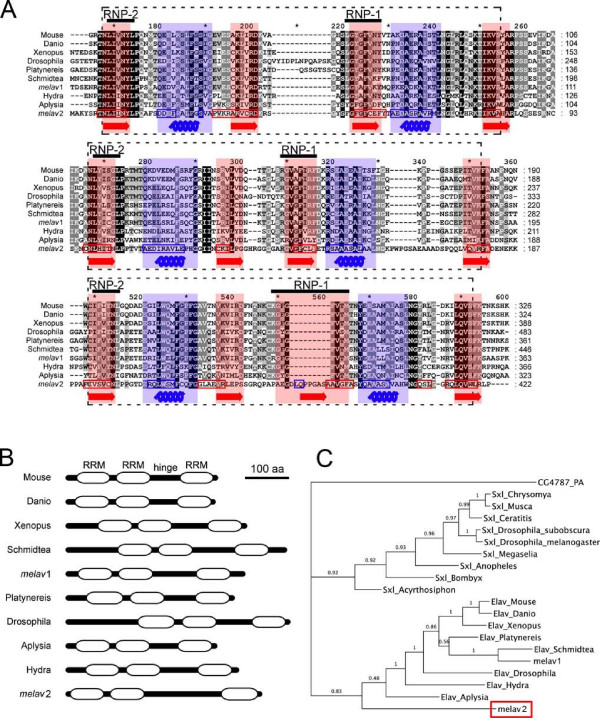
Figure 1.
Comparison of melav2 and other elav family genes. (A) Alignments of the melav2 amino acid sequence with other elav-like genes from different organisms. Three RNA recognition motifs (RRM) are present in all elav-like genes (each indicated by a dashed line box). Each RRM has two conserved sequences RNP-1 and RNP-2. Structurally, RRM consists of two alpha helixes (highlighted in blue and blue coil) and four beta sheets (highlighted in red and red arrow). The predicted corresponding sequence of melav2 is shown in blue and red boxes, respectively. (B) Predicted domain structure by the SMART program. Each box indicates an RRM. Melav2 had a similar gene structure that is similar to other elav family genes, namely three RRMs with a hinge region between the second and the third RRM. (C) Phylogenetic tree of melav2, other elav genes, and sxl genes from different organisms. Melav2 was categorized into the elav gene family. Numbers show the Bayesian posterior probability (with values > 0.95 representing good nodal support). For accession numbers see materials and methods. Mouse: Mus musculus, Xenopus: Xenopus laevis, Danio: Danio rerio, Drosophila: Drosophila melanogaster, Aplysia: Aplysia californica, Platynereis: Platynereis dumerilii, Schmidtea: Schmidtea mediterranea, Hydra: Hydra magnipapillata, melav1: another elav-like gene from M. lignano (see the discussion), Musca: Musca domestica, Ceratitis: Ceratitis capitata, Chrysomya: Chrysomya rufifacies, Megaselia: Megaselia scalaris, Anopheles: Anopheles gambiae, Acyrthosiphon: Acyrthosiphon pisum, Bombyx: Bombyx mori, CG4787-PA: a protein from D. melanogaster as an outgroup.