Figure 1. Algorithmic overview.
(A) A physical network model with nodes representing proteins and edges representing protein-DNA interactions. The sign of an interaction is denoted by its arrow type: regular (activating) or cut (suppressing). Note that the network is not sign-consistent since for example, 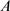 is linked to
is linked to 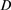 by two paths with different aggregate signs. (B) A functional network generated by the physical network (every knockout effect is explained by at least one path in the physical network, see Methods) with edges representing knockout effects and nodes representing the respective genes. The sign of a functional edge is denoted by its arrow type: regular (down-regulation) or cut (up-regulation). (C) The sign-linear algorithm. The functional network is translated into a set of Boolean equations. One optimal solution for the equations is setting
by two paths with different aggregate signs. (B) A functional network generated by the physical network (every knockout effect is explained by at least one path in the physical network, see Methods) with edges representing knockout effects and nodes representing the respective genes. The sign of a functional edge is denoted by its arrow type: regular (down-regulation) or cut (up-regulation). (C) The sign-linear algorithm. The functional network is translated into a set of Boolean equations. One optimal solution for the equations is setting  to
to  and the rest to
and the rest to  , satisfying all equations (green frame, bottom) but one (purple frame, top). The ensuing partition into two groups is depicted with edges corresponding to functional relations between groups. This partition can be used for predicting new knockout effects. (D) The sign-clustering algorithm. For each pair of nodes the presented
, satisfying all equations (green frame, bottom) but one (purple frame, top). The ensuing partition into two groups is depicted with edges corresponding to functional relations between groups. This partition can be used for predicting new knockout effects. (D) The sign-clustering algorithm. For each pair of nodes the presented  -value reflects their similarity in the functional network. A partition into clusters using a cutoff of
-value reflects their similarity in the functional network. A partition into clusters using a cutoff of  is depicted with edges defined as in panel C. This partition refines the one obtained by the sign-linear algorithm (3 groups instead of 2), correctly modeling all the knockout effects.
is depicted with edges defined as in panel C. This partition refines the one obtained by the sign-linear algorithm (3 groups instead of 2), correctly modeling all the knockout effects.

