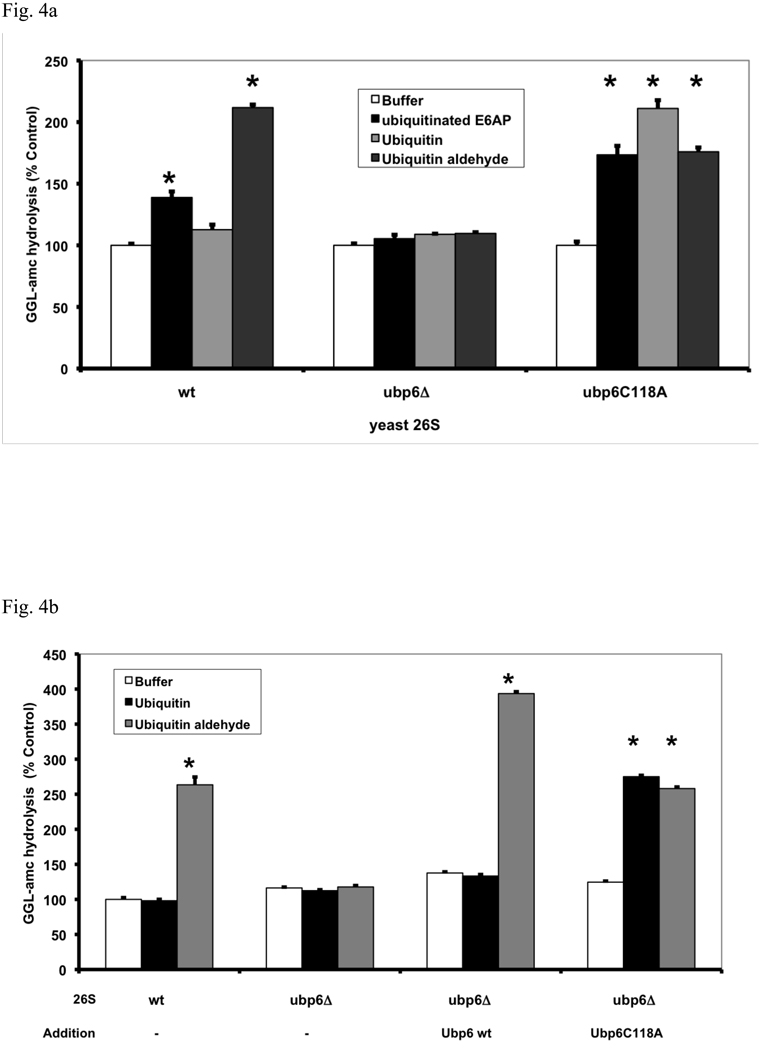
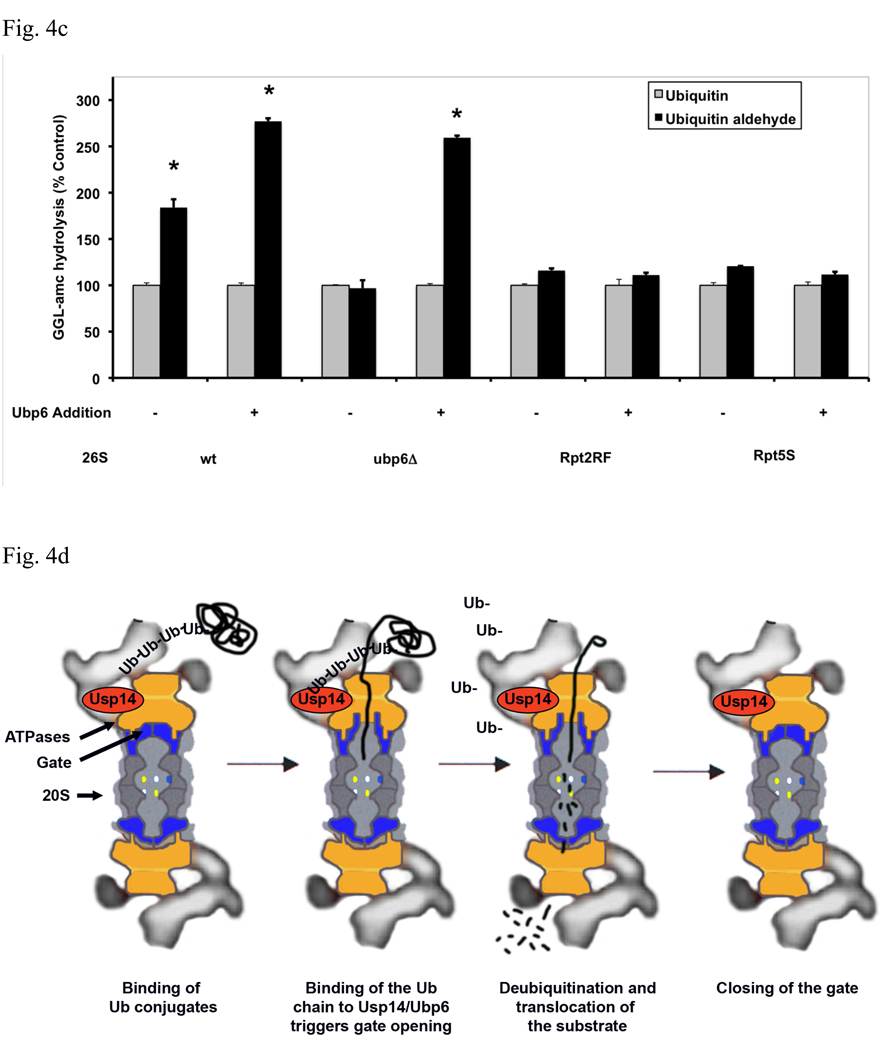
Fig. 4. Gate opening by Ub conjugates or Ub aldehyde requires Ubp6 protein in the proteasome but not its deubiquitinating activity.
(a) Ubiquitinated E6AP and Ub aldehyde stimulate activity of 26S proteasomes purified from wt yeast and strains expressing the UbpC118A active site mutant, but not from the ubp6 deletion mutant. The activity of untreated wild type 26S proteasomes was taken as 100%. The asterisk indicates P< 0.05. The stimulation by Ub aldehyde in wt yeast 26S proteasomes was consistently greater than by ubiquitinated E6AP. A large stimulation by Ub conjugates and free Ub was seen only in the enzymatically inactive Ubp6C188A mutant. (b) Reconstitution of stimulation (seen in Fig. 4a) was observed in Ubp6-deleted proteasomes after addition of purified wt or C188A mutant Ubp6. 26S proteasomes (5 nM) purified form ubp6 deletion strains were supplemented with wt Ubp6 or Ubp6C118A (20 nM). Peptide hydrolysis was monitored after the addition of Ub or Ub aldehyde. Wt and ubp6Δ proteasomes were used as positive and negative controls for stimulation by Ub aldehyde. The asterisk indicates P< 0.05. (c) Nucleotide binding to the ATPase subunits Rpt2 or Rpt5 is required for stimulation of 26S proteasomes by Ub aldehyde. Peptide hydrolysis was monitored after the addition of Ub or Ub aldehyde, no increase was observed in the Rpt2RF and Rpt5S proteasomes carrying mutations in their ATP-binding motif. 26S particles purified from wt and ubp6Δ strains were used as positive and negative controls for stimulation with Ub aldehyde. Addition of recombinant Ubp6 restores stimulation by Ub aldehyde in ubp6Δ, but not in Rpt2RF and Rpt5S proteasomes. The asterisk indicates P< 0.05. (d) Summary: The binding of Ub conjugates to Usp14 induces gate opening in the 26S proteasome. A cross section of the 26S proteasome shows binding of a ubiquitinated substrate to the 19S regulatory particle. Subsequently, the Ub chain interacts with Usp14, and during its cleavage, it induces maximal gate opening in the 20S particle. The Ub chain is disassembled, and the substrate is translocated into the 20S and hydrolyzed.