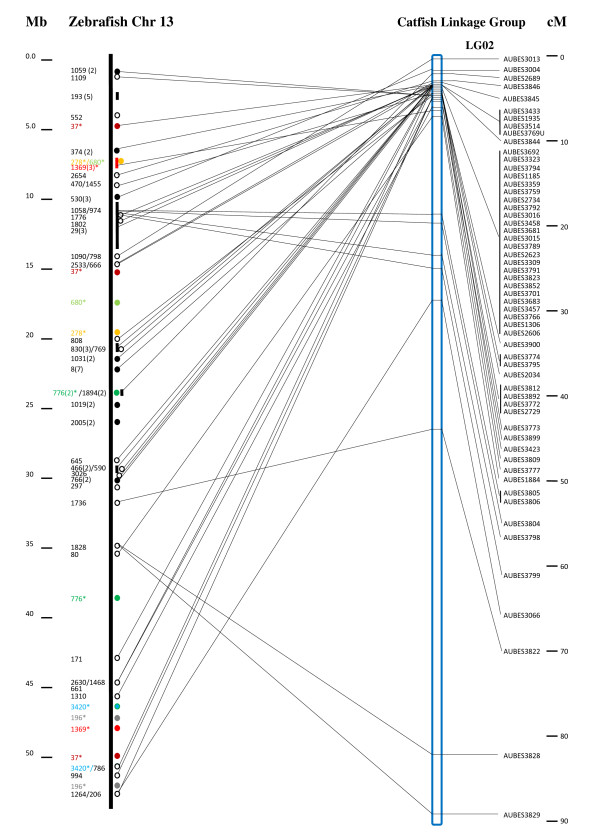
Figure 6.
Scaffolds of conserved syntenic regions between the catfish and zebrafish genomes. Scaffolds of conserved syntenic regions were established by genetic linkage mapping of BAC contig-associated microsatellites. The zebrafish chromosome 13 (chr 13) is presented with its base positions on the far left in million base pairs; The second column of the numbers are catfish BAC contig numbers [41], with the identified syntenic regions shown immediately right of the chromosome bar. The numbers in the parenthesis are the number of conserved sequences; the circles and bars represent relatively short and long conserved syntenic regions; the asterisks represent duplicated syntenic regions with color coding to facilitate the visualization of duplicated regions, the same way as described under Figure 1 legend, except that the open circles represent conserved sequences coming from non-gene sequences while the solid circles represent conserved gene sequences. Microsatellites from the BAC contigs were genetically mapped to linkage groups as shown on the right, with the names of microsatellites being labeled on the second most right, e.g., AUBES1884. The positional relationship of the conserved syntenies on the zebrafish genome sequence and within the catfish linkage group is indicated by thin lines linking the zebrafish chromosome and the catfish linkage group positions. The positions of markers within the linkage group are shown on the furthest right in centi-Morgans.