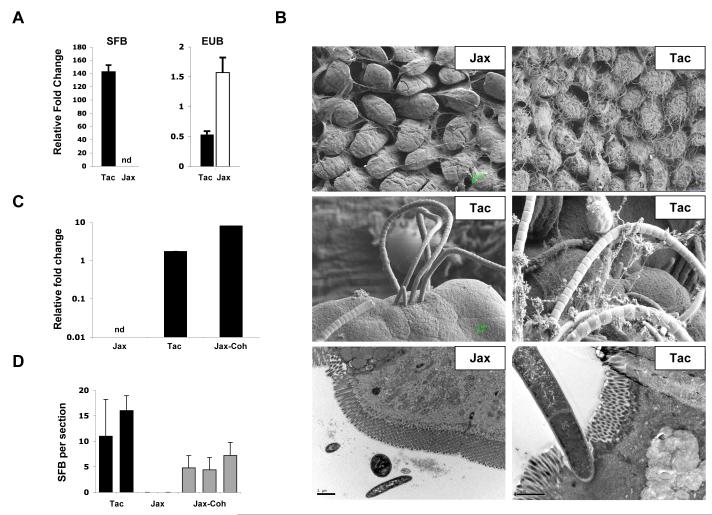
Figure 2. Segmented Filamentous Bacteria (SFB) in the intestinal tract of Th17 cell-sufficient and Th17 cell-deficient mice.
A. Quantitative PCR (qPCR) analysis of SFB and total bacterial (EUB) 16S rRNA genes in mouse feces from Taconic (Tac) and Jackson (Jax) B6 mice. Genomic DNA was isolated from combined fecal pellets from 4 animals from each strain. The experiment was repeated numerous times with similar results.
B. Scanning (SEM) and transmission (TEM) electron microscopy of terminal ileum of 8 week-old Jackson (Jax) and Taconic (Tac) C57BL/6 mice housed under similar conditions and diet for at least one week. Note the presence of long filamentous bacteria with SFB morphology in Taconic, but not Jackson mice
C. qPCR analysis for SFB presence in Jackson B6 mice after 14 days of co-housing with Taconic B6 mice (Jax-Coh). Genomic DNA was isolated from pooled feces from 3-4 mice per group.
D. SFB colonization of terminal ileum of Jackson B6 mice after 14 days of co-housing with Taconic B6 mice (Jax-Coh). Toluidine-blue sections were prepared from 0.5 cm piece of the terminal ileum as described in Methods and examined by light microscopy. Adherent bacteria with SFB morphology were counted in 4-5 sections from each sample. Each column represents a separate animal.