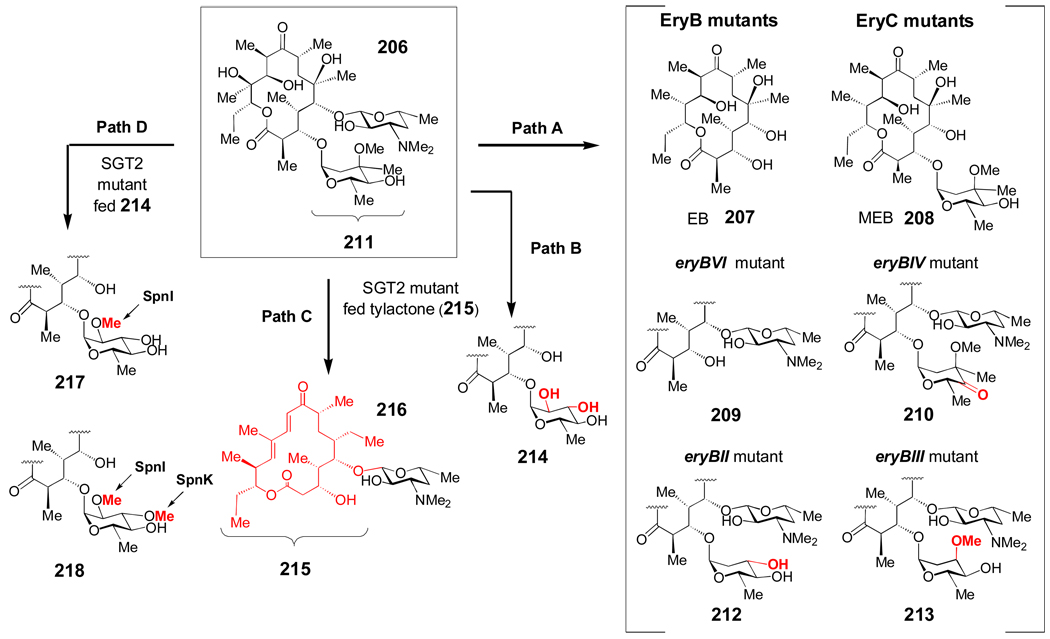
Figure 7. Engineering erythromycin derivatives in Saccharopolyspora erythraea with gene disruption, heterologous expression, and feeding experiments.
Path A – Disruption of individual l-mycarose (eryB) or d-desosamine (eryC) biosynthetic genes afforded 207–210, 212, and 213. Path B – Heterologous expression of the oleandrosyltransferase (OleG2) from Streptomyces antibioticus in an S. erythraea mutant lacking the endogenous mycarosyltransferase (EryBV). Path C – Expression of the mycaminosyl transferase (TylM2) from the tylosin pathway of Streptomyces fradiae in a triple S. erythraea mutant (SGT2) deficient in desosaminyltransfer (eryCIII−), mycarosyltransfer (eryBV−), and polyketide synthesis (eryA−), generated 216 when the strain was fed tylactone (215). Path D – Novel erythromycin derivates (217 and 218) generated when O-methyltransferase genes (spnI and spnK) from Saccharopolyspora spinosa were heterologously expressed in the S. erythraea SGT2 mutant.