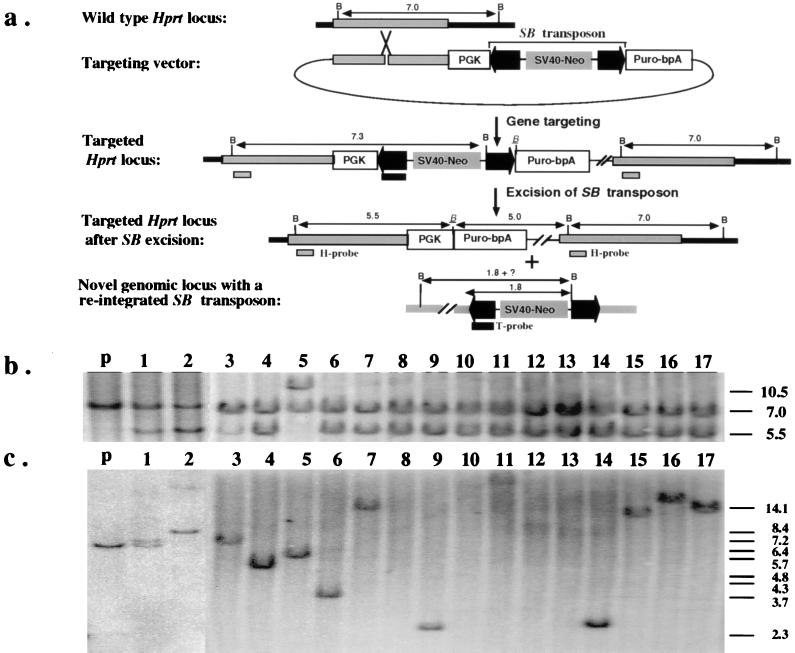
Figure 2.
(a) Introduction of a single copy of the excision testing construct into the Hprt locus by gene targeting. The region of homology for the insertional-type targeting vector is indicated as gray bars. The PGK promoter and the coding sequence of the puromycin-resistance gene are shown as open boxes. The IR/DR sequences of the SB transposon are highlighted as dark arrowheads. A complete, nonautonomous SB element also is indicated in the targeting vector. The positions of the Hprt- (H) and transposon-specific (T) probes that detect excision and reinsertion events, respectively, also are indicated. All relevant BamHI sites are shown as “B” except the one that is adjacent to the excision site, which is italicized and underlined. Notice that the SB element contains a unique BamHI site approximately 1.8 kb from the end that hybridizes to the T probe. (b) Southern hybridization analysis with the H probe confirms the excision of the SB element from the testing construct. The lower intensity of the 5.5-kb fragment compared with the 7.0-kb fragment in clone 1 and clone 3 is a result of contamination with parental cells as the result of cross-feeding. The parental line has a predicted doublet consisting of a 7.0-kb and a 7.3-kb fragment. (c) Southern hybridization analysis with the T probe confirms reintegration of the excised SB element. The sizes of the bands in lanes 1, 2, 3, 4, 5, 6, 9, 11, 14, 15, 16, and 17, are variable but are all longer than 2.3 kb. No hybridization was detected in lanes 8, 10, 12, and 13. The parental fragment detected in clones 1 and 3 is due to parental cell contamination. The size marker is BstEII-digested λ-phage DNA.