Table 2.
Parameter settings
| parameter | R5 virus | X4 virus |
|---|---|---|
| number of strains | nR5 | nX4 |
| viral growth rate ri |
r (2d-1) |
r(1 + αz) ((2 + z)d-1) |
| effect of specific immune response pi |
pR5 (2d-1) |
pX4 >pR5 (20d-1) |
| effect of cross-reactive immunity qi |
q (1.86d-1) |
|
| stimulation of specific and cross-reactive immunity ci, ki |
c, k (0.1d-1V L-1) |
|
| natural and virus induced depletion of immune response b, ui |
b, u (0.02d-1, 0.1d-1V L-1) |
|
| probability to generate a new strain pm per day and viral load |
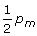 (0.005d-1V L-1) |
|
Parameter settings for R5 and X4 viruses, the values chosen in the simulations are given in brackets in units per day (d) and viral load (VL), due to the close genetic neighbourhood of R5 and X4 viruses it is assumed that both mutants occur with the same probability 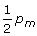 from the total viral population irrespective of its composition. Where direct biological interpretation is possible parameters have been chosen following [29,30], remaining degrees of freedom have been fixed to account for the appropriate dynamical regime defined by equation (4). Note that the growth rate of X4 viruses is assumed to increase proportional to the immune activation measured by the cross reactive immunity z with α = 0.5.
from the total viral population irrespective of its composition. Where direct biological interpretation is possible parameters have been chosen following [29,30], remaining degrees of freedom have been fixed to account for the appropriate dynamical regime defined by equation (4). Note that the growth rate of X4 viruses is assumed to increase proportional to the immune activation measured by the cross reactive immunity z with α = 0.5.
