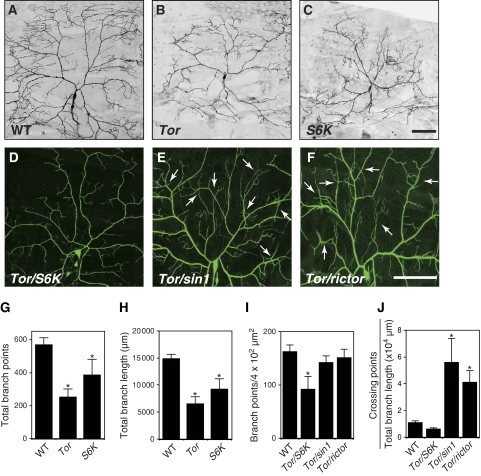
Figure 3.
TORC1 and TORC2 regulate dendritic growth/branching and tiling, respectively. (A–C) Tor and S6K MARCM clones are defective in both dendritic arborization and branching. MARCM clones of (A) wild-type (WT), (B) TorΔP, and (C) S6Kl−1 are shown. Bar represents 50 μm. Clone genotypes: (A) hsFLP, elavGal4, UAS-mCD8-GFP/+; FRT40A, (B) hsFLP, elavGal4, UAS-mCD8-GFP/+; FRT40A, TorΔP; (C) hsFLP, elavGal4, UAS-mCD8-GFP/+; +/+; FRT82B, S6Kl−1. (D–F) Live images of ddaC dendrites visualized by the ppk-EGFP reporter in third instar larvae trans-heterozygous for TorΔP and S6Kl−1 (D), for TorΔP and sin1PBac (E), and for TorΔP and rictorΔ2 (F). Arrows in (E) and (F) indicate the crossing points of the dendritic branches. Genotypes: (D) TorΔP/+; S6Kl−1, ppk-EGFP/ppk-EGFP, (E) TorΔP/sin1PBac; ppk-EGFP/ppk-EGFP, and (F) rictorΔ2/+; TorΔP/+; ppk-EGFP/ppk-EGFP. (G, H) Quantification of the total branch points (G) and the branch length (H) of MARCM clones. Error bars indicate the mean±s.d. (WT, n=6; Tor, n=11; S6K, n=5), *P<0.01 relative to WT controls (Student's t-test). (I, J) Quantification of the branch points (I) and the crossing points (J) per μm2 (4 × 104) of the dendritic branches at the third instar larval stage in ddaC neurons. Error bars indicate the mean±s.d. (n=15), *P<0.01 (Student's t-test).