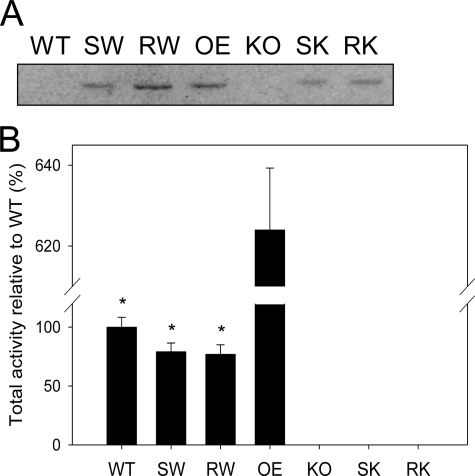
FIGURE 2.
Expression of site-directed AtFAAH mutants in Arabidopsis and evaluation of their enzyme activities. A, Western blot analysis. Arabidopsis wild-type (WT) and transgenic lines (SW, RW, OE, KO, SK, and RK) were grown for 10 days as described under ”Experimental Procedures.“ Total proteins (200 μg) from each seedling homogenates were analyzed by Western blot. All AtFAAH proteins of overexpressors (SW, RW, OE, SK, and RK) were immunolocalized at the position expected (∼66.1 kDa). SW, S281A/S282A mutant expressed in wild-type background; RW, R307A mutant expressed in wild-type; OE, native AtFAAH overexpressor; KO, Atfaah T-DNA insertion knock-out; SK, S281A/S282A mutant expressed in Atfaah knock-out background; RK, R307A mutant expressed in Atfaah knock-out. B, NAE hydrolysis activities. Total proteins (50 μg) from each of 10-day-old seedling homogenates were reacted with [1-14C]NAE16:0. Total activities (micromole/h) were measured based on radioactivity of the product formed. All AtFAAH overexpressors with site-directed mutation (SW, RW, SK, and RK) exhibited significantly lower NAE hydrolase activity than the native AtFAAH overexpressor (OE). The error bars represent S.D. from triplicate measurements. Asterisks indicate a significant difference (p < 0.0001) compared with OE, which was determined by Student's t test.