Abstract
In Escherichia coli, regulatory inactivation of the replication initiator DnaA occurs after initiation as a result of hydrolysis of bound ATP to ADP, but it has been unknown how DnaA is controlled to coordinate cell growth and chromosomal replication in Gram-positive bacteria such as Staphylococcus aureus. This study examined the roles of ATP binding and its hydrolysis in the regulation of the S. aureus DnaA activity. In vitro, S. aureus DnaA melted S. aureus oriC in the presence of ATP but not ADP by a mechanism independent of ATP hydrolysis. Unlike E. coli DnaA, binding of ADP to S. aureus DnaA was unstable. As a result, at physiological concentrations of ATP, ADP bound to S. aureus DnaA was rapidly exchanged for ATP, thereby regenerating the ability of DnaA to form the open complex in vitro. Therefore, we examined whether formation of ADP-DnaA participates in suppression of replication initiation in vivo. Induction of the R318H mutant of the AAA+ sensor 2 protein, which has decreased intrinsic ATPase activity, caused over-initiation of chromosome replication in S. aureus, suggesting that formation of ADP-DnaA suppresses the initiation step in S. aureus. Together with the biochemical features of S. aureus DnaA, the weak ability to convert ATP-DnaA into ADP-DnaA and the instability of ADP-DnaA, these results suggest that there may be unidentified system(s) for reducing the cellular ratio of ATP-DnaA to ADP-DnaA in S. aureus and thereby delaying the re-initiation of DNA replication.
Introduction
The initiation step of chromosome replication is an important set point for coordinating cell growth and chromosomal replication, and it is tightly regulated so that it takes place only once per cell cycle. DNA replication in bacteria starts when the cell mass reaches an appropriate size and at the appropriate time, which is controlled in part by the quantity and activity of initiator DnaA protein (1–3). In Escherichia coli, DnaA binds to 9-mer DnaA boxes in oriC, the origin of chromosome replication, and melts the duplex at AT-rich 13-mers adjacent to oriC. DnaA then directs loading of the DnaB replicative helicase, which depends on the DnaC helicase loader and priming by the DnaG primase. This is followed by the synthesis of DNA by the DNA polymerase III holoenzyme (4).
DnaA is composed of four domains: N-terminal domains I and II are involved in DnaA oligomerization and DnaA-DnaB interaction, and they show a diversity of sequence among bacteria. Domain III contains AAA+ motifs, and the C-terminal domain IV mediates DNA binding (3, 5). Biochemical and genetic studies in E. coli have revealed adenine nucleotide binding-mediated control of the DnaA activity (6–11). Specifically, ATP-bound DnaA actively promotes replication initiation, whereas the ADP-bound form is inert (6, 7). ATP promotes the self-assembly of DnaA at replication origins via a conformational change to form a right-handed helical filament (8, 9). In E. coli, 70% to 80% of DnaA exists as the inactive ADP-bound form, and active ATP-DnaA levels increase during the initiation of chromosome replication (10). The ratio of ATP-DnaA to ADP-DnaA is regulated by de novo synthesis of the ATP-bound DnaA, hydrolysis of bound ATP to ADP by the intrinsic ATPase activity of DnaA, the stimulatory factor RIDA (regulatory inactivation of DnaA),6 and the exchange of bound ADP for ATP (10, 12). RIDA requires a β clamp of DNA polymerase III and Hda protein and negatively regulates the initiation step of DNA replication by promoting hydrolysis of bound ATP on DnaA, which is coupled to the progression of chromosome replication (13–18). The factors to promote the exchange of bound ADP for ATP on DnaA include fluid membranes with acidic phospholipids as well as a specific DNA sequence containing DnaA boxes (19–22). DnaA acts as a transcription factor and autoregulates its expression (23–26), and its activity is affected by nucleotide binding (27, 28).
Comparison of replication-related gene sequences suggests that there are different mechanisms for controlling initiation in Gram-positive bacteria with low GC content such as Staphylococcus aureus and Bacillus subtilis than in Gram-negative bacteria such as E. coli. Specifically, Gram-negative bacteria related to E. coli contain Dam methylase and SeqA (29–31), which inactivate oriC and the dnaA promoter immediately after the initiation of DNA replication, whereas these proteins are absent in Gram-positive bacteria. Conversely, Gram-positive bacteria contain DnaB and DnaD proteins, which are involved in the initiation step of DNA replication (32–37). Gram-positive bacteria also express an additionally essential catalytic subunit of DNA polymerase III (38, 39). Furthermore, two regulators of DnaA in E. coli, Hda described above and DiaA (40), are not present in S. aureus and B. subtilis. Instead, these Gram-positive bacteria contain YabA, which negatively regulates the initiation step of DNA replication by binding to DnaA and DnaN (41–43). Although DnaA is widely conserved in prokaryotes and is essential in both Gram-negative and Gram-positive bacteria, DnaA of B. subtilis and S. aureus is not interchangeable with E. coli DnaA (3, 44). Therefore, DnaA differs in its function to control the replication initiation in each bacterium, but DnaA must function as an initiator in both. However, there is little information on how DnaA is regulated in bacteria other than E. coli.
The present study showed that ADP dissociates much faster from purified S. aureus DnaA than from E. coli DnaA, so that ADP-bound S. aureus DnaA was rapidly converted to the ATP-bound form in vitro. This raises a question whether DnaA is really inactivated by ADP binding in S. aureus. To explore this question further, we examined the similarities and differences in initiation control of chromosome replication by S. aureus DnaA. In vitro, S. aureus DnaA mediated open complex formation at oriC in an ATP-dependent manner, although hydrolysis of ATP was not required. In S. aureus cells, induced expression of an S. aureus DnaA R318H mutant protein, which has greatly reduced intrinsic ATPase activity, caused excessive initiation. These results suggest that, as in E. coli, ADP-DnaA negatively regulates the initiation step of chromosome replication and DnaA concentration alone is not the only factor determining the timing of the initiation in S. aureus. The results also imply that S. aureus possesses a system for maintaining the ADP-bound form of DnaA.
EXPERIMENTAL PROCEDURES
Bacteria, Plasmids, and Culture Conditions
E. coli and S. aureus cells were grown in Luria-Bertani (LB) medium (1% Tryptone (Difco), 0.5% yeast extract (Difco), and 1% NaCl) with 25 μg/ml thymine, 50 μg/ml ampicillin, 25 μg/ml kanamycin, 12.5 μg/ml chloramphenicol, or 10 μg/ml erythromycin as required. S. aureus strains RN4220, NI8 (thyA19::pSF151kan as RN4220), and NM1003 (Ω[sa0009-sa0010::pMutinT3erm] as NI8) were described previously (44). E. coli strain KA450 (ΔoriC1071::Tn10, dnaA17(Am), rnhA199(Am)) or KP7364 (ΔdnaA::spec rnhA::kan) was used for construction of oriC plasmids or plasmids containing DnaA from S. aureus, B. subtilis, Streptococcus pyogenes, and E. coli. E. coli JM109 (recA1) was used as a host during construction of other plasmids and to prepare a monomer form of pCK206 (oriC) DNA. The shuttle vectors between E. coli and S. aureus were pND50 (45), pKE515 (37), and their derivatives.
Construction of oriC Plasmids
To construct the pCK206 oriC plasmid, the dnaA-coding region with the proximal upstream and downstream region (3.5 kbp), which included rnpA, rpmH, dnaA, and the 5′-region of dnaN, was amplified by PCR with primers 5′-GGAATTCCCCCTACCCTATCCTTACTTAATCTT-3′ and 5′-TTGTATAAGCCAGTTCACACCAGTTA-3′ and using an RN4220 genomic DNA as a template. The amplified fragment was cloned into the SmaI site of the E. coli pUC-derived plasmid pCK20 (46). The plasmid obtained was digested with EcoRV and PstI, blunt-ended, and self-ligated to truncate dnaA and delete its upstream region. The resulting plasmid was named pCK206, and it contained the 3′-region of dnaA to the 5′-region of dnaN as oriC.
Construction of Plasmids for Overexpressing S. aureus DnaA
A wild-type S. aureus DnaA-overproducing plasmid, pBAD-dnaA, was generated as follows. The coding region of the S. aureus dnaA gene (1.4 kbp) was amplified by PCR using primers 5′-GGGAATTCACCATGTCGGAAAAAGAAATTTGGG-3′ (EcoRI site underlined) and 5′-GGCTGCAGTTATACATTTCTTATTTCTTTTTC-3′ (PstI site underlined) and pHYdnaA (44) as a template. The resulting fragment was subcloned into the pGEM-T vector (Promega), excised with EcoRI and PstI, and cloned into the EcoRI and PstI site of pBAD24, which contains the arabinose promoter and operator (47), resulting in pBAD-dnaA. To construct pBAD-R318H, the entire region of pBAD-dnaA was amplified by PCR with primers 5′-CATGAATTAGAAGGTGCATTAACACG-3′ and 5′-AATATTAGATTGAATTTGATTTGC-3′ (altered nucleotide underlined), which introduces an Arg to His substitution at residue 318 of S. aureus DnaA. After self-ligation, the resulting plasmid was introduced into and amplified in E. coli strain KA450, resulting in pBAD-R318H. Sequence analysis confirmed that the open reading frame (ORF) of DnaA in this plasmid lacked secondary mutations.
Construction of Atc-inducible Plasmids for Expressing DnaA in S. aureus
The DNA region (0.8 kbp) from pWH353 (48), which includes the tetR gene and a synthetic xyl-tet promoter-operator fusion, was amplified by PCR using primers 5′- GCCCAAGCTTTAAAATCGATAACTCGACAT-3′ and 5′-AGATCTGATATCAAGCTTATTTTAA-3′ (HindIII sites underlined). The amplified DNA fragment was digested with HindIII and ligated into HindIII-digested pND50, resulting in pNDX1. Next, the DNA fragment, including the S. aureus dnaA ORF along with the Shine-Dalgarno sequence (1.5 kbp), was amplified by PCR using primers 5′-CCGGAATTCGTGTATAACTTAAAAATTTAA-3′ (EcoRI site underlined) and 5′- GCCGGATCCAAAGTTTCCTACTTATACATT-3′ (BamHI site underlined). The amplified DNA fragment was ligated into SmaI site of the pNDX1, resulting in pNDX1-dnaA. Although pNDX1-dnaA contained a single tet regulatory element, we replaced it with double tet operators derived from pWH354, which enabled tighter repression (48). The synthetic xyl-tet-tet promoter-operator-operator region of pWH354 was excised with XhoI and PstI and then cloned into the XhoI and PstI site of pNDX1-dnaA. The resulting plasmid, pNDX2-dnaA, was used for Atc-inducible expression of DnaA in S. aureus. To construct pNDX2-R318H, the EcoRV fragment (0.4 kbp) of pBAD-R318H, which includes the internal DnaA protein region carrying the R318H mutation, was inserted into EcoRV-digested pNDX2-dnaA. The pNDX2 vector was prepared from pNDX2-dnaA by EcoRI digestion followed by self-ligation, which removed the cloned dnaA region.
Construction of Multicopy dnaN Plasmid
The multicopy dnaN plasmid, pSE-dnaN, was constructed as follows. The DNA region, including the S. aureus dnaN ORF along with the Shine-Dalgarno sequence (1.2 kbp), was amplified by PCR using primers 5′-AAGGTACCTATATAATTATATATAAACGACT-3′ (KpnI site underlined) and 5′-GTGCGGCCGCAATACTGTCATTTTCAATTTTT-3′ (NotI site underlined) and chromosomal DNA from S. aureus strain RN4220 as a template. The amplified product was digested with KpnI and NotI and then cloned into the KpnI and NotI sites of pSRP21, resulting in pSRP-dnaN. The pSRP21 plasmid was a pKE515-derived plasmid carrying the upstream region of the S. aureus hisS gene (0.3 kbp; amplified with primers 5′-GCCTCTAGAACATTGAACACAAGAAGAGGT-3′ and 5′-GCCGAATTCTCGGCAAACTATTTTATTTAA-3′) inserted into the SmaI site. Plasmid pSRP-dnaN was digested with BamHI and NotI, and then a 1.6-kbp fragment containing the hisS upstream region and the dnaN-coding region was cloned into BamHI/NotI-digested pKE516, resulting in pSE-dnaN. Plasmid pKE516 was a derivative of pKE515, wherein the chloramphenicol resistance gene of pKE515 was replaced with the erythromycin resistance gene.7
Purification of S. aureus DnaA Protein
S. aureus DnaA was purified as described previously (46) with some modifications. Briefly, lysates from E. coli strain KA450 harboring pBAD-dnaA were prepared and purified by Mono S column chromatography as described previously (46). DnaA-containing fractions were identified by ATP binding, pooled, diluted with an equal volume of buffer D (50 mm HEPES-KOH, pH 7.6, 1 mm EDTA, 2 mm dithiothreitol (DTT), 20% glycerol, 10 mm magnesium acetate, and 0.2 m potassium sulfate) containing 7 m guanidine hydrochloride and then centrifuged at 13,000 × g for 20 min at 4 °C. The supernatant was applied to a Superose 12 10/30 fast protein liquid chromatography column (Amersham Biosciences) equilibrated with buffer D, and DnaA protein was eluted with buffer D. Fractions showing ATP binding were pooled and used as purified S. aureus DnaA. S. aureus DnaAR318H protein was purified form E. coli KA450 harboring pBAD-R318H in the same way. In both cases, SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250 revealed a single band (see Fig. 3A). Protein concentrations were determined by the Bradford method using bovine serum albumin (BSA) as a standard.
FIGURE 3.
Characteristics of S. aureus DnaAR318H in vitro. A, wild-type or R318H DnaA (500 ng) was separated by SDS-PAGE (12.5%) and visualized with Coomassie Brilliant Blue R-250. B, ATP dissociation. Wild-type DnaA (closed circles) or R318H (open circles) (5 pmol) was preincubated with 1.0 μm [α-32P]ATP on ice and then mixed with 1 mm ATP and incubated at 37 °C. The amount of bound ATP was measured with a filter binding assay. Results shown are means ± S.E. of at least three independent experiments. C, ATPase activity. Wild-type DnaA (20 pmol, closed circles), R318H DnaA (20 pmol, open circles), or without DnaA (closed squares) was preincubated with 1.0 μm [α-32P]ATP on ice for 30 min and then incubated at 37 °C for the indicated time. Samples were separated by TLC, and the amount of ADP was determined using fluorography. Results are means ± S.E. of at least three independent experiments. D, open complex formation. pCK206 (100 fmol) was incubated with wild-type or R318H DnaA and digested with P1 nuclease as described in Fig. 2B.
Purification of B. subtilis DnaA and Streptococcus pyogenes DnaA Proteins
B. subtilis DnaA protein ORF was amplified with PCR using primers 5′-ACGTGCCGGACCATGGAAAATA-3′ (NcoI site underlined) 5′-CCCCCATGGATCCCCGGTCCTGCTATT-3′ (NcoI site underlined) and B. subtilis 168 genomic DNA as a template and cloned into the NcoI site of pBAD24 (41). The resulting plasmid, pBAD-BdnaA, was introduced into an E. coli strain KP7364, which lacks the E. coli dnaA gene (11). S. pyogenes DnaA protein ORF was amplified with PCR using primers 5′-TCCACAAGAAGGAATTCAGTATGACTGAA-3′ (EcoRI site underlined) 5′-ACGAGAGAATTCAAGACTAAATGG-3′ (EcoRI site underlined) and S. pyogenes SSI-9 genomic DNA as a template and cloned into the EcoRI site of pBAD24. The resulting plasmid, pBAD-SpydnaA, was introduced into an E. coli strain KA450. Induction of B. subtilis DnaA and S. pyogenes DnaA proteins was carried out at 16 °C, and purifications of these proteins was followed by the procedure for S. aureus DnaA as descried above. DNA sequences of dnaA ORFs in overexpressing plasmids were confirmed by DNA sequencing.
Purification of S. aureus HU Protein
S. aureus HU protein consists of a homodimer. The S. aureus HU protein ORF was amplified with PCR using primers 5′-AGGAGGTGAATGTCTCATGAACAAA-3′ (BspHI site underlined) 5′-TTTTGAAAGCTTAAACAATGAATGTTCC-3′ (HindIII site underlined) and cloned into the NcoI and HindIII sites of pBAD24. The resulting plasmid, pBAD-HU, was introduced into E. coli strain RI-07, which lacks the hupA and hupB genes (49). RI-07 harboring pBAD-HU was cultured in 1 liter of LB medium containing 100 μg/ml ampicillin at 37 °C until the A600 reached 0.2, after which the culture was adjusted to 0.2% (w/v) l-(+)arabinose. After a 2-h induction, cells were harvested by centrifugation, washed with 0.9% NaCl, suspended in 1 ml of buffer A (20 mm Tris-HCl, pH 7.5, 5 mm EDTA, 0.5 mm DTT, 10% glycerol, and 50 mm NaCl) per milliliter of wet cell weight, and frozen in liquid N2. The frozen cell paste was thawed on ice and adjusted to 0.4 mg/ml lysozyme and 30 mm spermidine. After 30-min incubation on ice for cell lysis, samples were again frozen, thawed on ice, and then sonicated four times for 10 s using a Branson Sonifier 450 with an ultramicrotip at an output setting of 3. The resulting homogenate was centrifuged at 35 krpm for 30 min at 4 °C using a Beckman 80Ti rotor, and the cleared lysates obtained were loaded onto a DNA-cellulose column pre-equilibrated with buffer A. The column was then washed, and proteins were eluted with a linear gradient from 50 mm to 2 m NaCl in buffer A. The presence of HU protein was determined by SDS-PAGE, and a fraction of peak, which represented a single band stained with Coomassie Blue, was used as purified HU protein. The purified HU protein also had the ability to cause constrained supercoiling of pUC119 form I DNA in the presence of calf thymus DNA topoisomerase I (50). The protein concentration was determined by the Lowry method using BSA as a standard. Proteins were stored at −80 °C.
ATP and ADP Binding by DnaA
A filter binding assay was used to measure ATP binding by DnaA as described previously (46), except that the magnesium concentration was increased to 10 mm. The same conditions were applied for all DnaA proteins form S. aureus, B. subtilis, S. pyogenes, and E. coli. Typical reactions were performed on ice in a 50-μl solution of 40 mm HEPES-KOH, pH 7.6, 100 mm potassium glutamate, 10 mm magnesium acetate, 0.5 mm EDTA, 1 mm DTT, 50 μg/ml BSA, 10% sucrose, 1 μm [α-32P]ATP (40,000 cpm/pmol), or [3H]ADP (11,400 cpm/pmol), and purified S. aureus DnaA proteins (50–250 ng). DTT was added just before initiating the reaction. Kd values were determined by incubating 5 pmol of purified S. aureus DnaA proteins for 3 h on ice with ATP or ADP and in a volume of 0.5 ml or 5 ml, respectively. ATP and ADP binding in dissociation and exchange reactions were also detected by the filter binding assay.
ATPase Activity of S. aureus DnaA
DnaA protein (20 pmol) was preincubated on ice for 30 min with pCK206 (100 ng) and 1 μm [α-32P]ATP (40,000 cpm/pmol) in 20 μl of a reaction buffer containing 40 mm HEPES-KOH, pH 7.6, 100 mm potassium glutamate, 1 mm magnesium acetate, 0.5 mm EDTA, 1 mm DTT, 50 μg/ml BSA, and 10% sucrose and then incubated at 37 °C. A portion (2 μl) of samples was placed on ice to stop the reaction, after which 1 μl was spotted onto a silica-gel TLC plate. The plate was developed with 1 m formic acid containing 0.6 m lithium chloride and then exposed to a BAS MS2025 imaging plate (Fuji). The plates were analyzed with a BAS-1800II bioimaging analyzer (Fuji).
P1 Nuclease Assay
S. aureus DnaA was preincubated at 37 °C for 20 min in a reaction buffer (10 μl) containing 40 mm HEPES-KOH, pH 7.6, 100 mm potassium glutamate, 10 mm magnesium acetate, 0.5 mm EDTA, 1 mm DTT, 50 μg/ml BSA, 10% sucrose, and 5 mm ATP or ADP. In another tube, pCK206 oriC DNA (100 fmol) was also preincubated at 37 °C for 10 min in the same reaction buffer (40 μl) containing 100 ng of S. aureus HU protein. Next, DnaA was added to the pCK206 DNA, and the mixture was incubated for an additional 10 min at 37 °C. P1 nuclease (Roche Applied Science, 0.3 unit; or MP Biochemicals, 10 units) was then added, and the mixture was incubated for 25 s at 37 °C. The nuclease reaction was stopped by the addition of 50 μl of SDS-EDTA (0.2% SDS and 40 mm EDTA), and the proteins were removed by phenol/chloroform extraction. DNA in the samples was recovered by ethanol precipitation, separated by 1% agarose gel electrophoresis, and stained with ethidium bromide.
Western Blotting for S. aureus DnaA
To estimate the level of DnaA induction in vivo, S. aureus NM1003 cells harboring an empty pNDX2 vector, pNDX2-dnaA, or pNDX2-R318H were grown at 37 °C until the A600 reached 0.1. The cells were then treated for 1 h with 0, 1, or 10 ng/ml Atc. Proteins were extracted from the cells, determined by the Lowry method with BSA as a standard, and then analyzed by Western blotting for S. aureus DnaA as described previously (44).
Southern Hybridization
Southern blotting to determine the origin-to-terminus ratio was performed as described previously (37).
Flow Cytometry
Flow cytometric analysis to determine chromosome equivalents per cell was performed as previously described (36).
Measurement of DNA Synthesis
DNA synthesis in S. aureus cells was determined by [methyl-3H]thymine incorporation into acid-insoluble material as previously described (44).
RESULTS
Rapid Exchange of Bound ADP for ATP by S. aureus DnaA
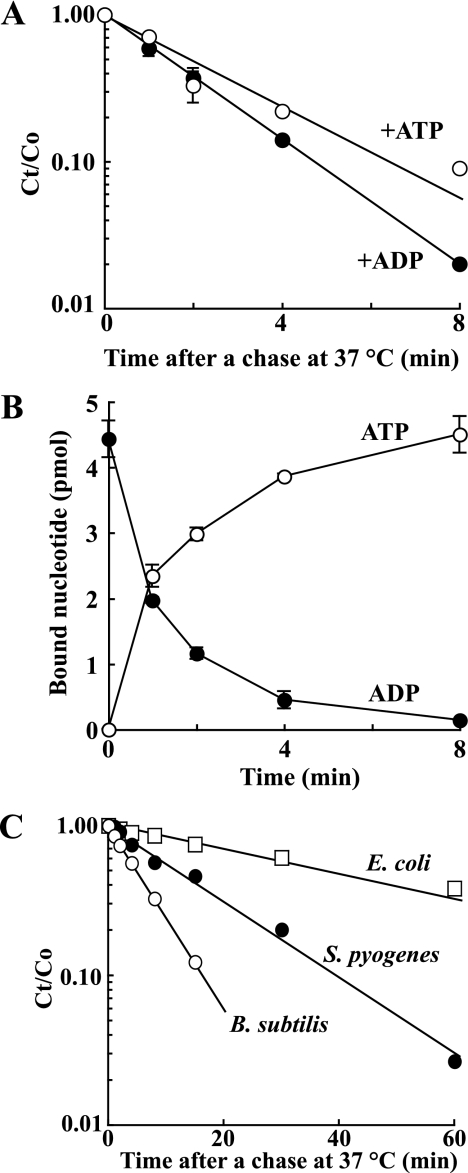
S. aureus DnaA binds ATP and ADP with high affinity in vitro (46). Here, we measured the dissociation rate of the [3H]ADP-DnaA complex in the presence of excess concentrations of ATP or ADP. The half-life of ADP-DnaA was 15 min on ice (data not shown) and ∼1.5 min at 37 °C (Fig. 1A). In agreement with this, when [3H]ADP-DnaA was chased with [α-32P]ATP at 37 °C, the half-life for exchange was 1 min (Fig. 1B). This is quite different from E. coli ADP-DnaA, which requires ∼45 min to exchange half of ADP-DnaA with ATP-DnaA under similar conditions at 38 °C (6), although the affinities to adenine nucleotides are compatible between S. aureus and E. coli DnaA. For E. coli DnaA, a low concentration of Mg2+ (0.2 mm) or a higher concentration of ATP than Mg2+ destabilizes ADP-DnaA (51). Therefore, we used a high Mg2+ (10 mm) concentration in our experiments. Regardless, we did not find conditions that stabilized S. aureus ADP-DnaA. Also, DNA with or without oriC did not stabilize S. aureus ATP-DnaA (data not shown). We also observed this instability of nucleotide binding for purified B. subtilis DnaA protein (Fig. 1C), in which the half-life of ATP-DnaA from B. subtilis was 5 min at 37 °C. This result suggests that the rapid nucleotide dissociation rate for S. aureus DnaA is not restricted in this bacterium. Meanwhile, the half-life of ATP-DnaA from Streptococcus pyogenes was 12 min at 37 °C (Fig. 1C), which was an intermediate value between 1.5 min of S. aureus ATP-DnaA and 41 min of E. coli ATP-DnaA.
FIGURE 1.
ADP binding by S. aureus DnaA is unstable. A, dissociation of ADP-DnaA. S. aureus DnaA (5 pmol) was preincubated with 1 μm [3H]ADP on ice and then mixed with 1 mm ATP (open circle) or ADP (closed circle) and incubated at 37 °C. B, exchange of bound ADP for ATP by DnaA. S. aureus DnaA (30 pmol and 10 μl) was preincubated with 1 μm [3H]ADP at 37 °C for 9 min and then mixed with 40 μl of a solution containing 12.5 μm [α-32P]ATP and incubated at 37 °C for the indicated time. The levels of ADP-bound (closed circle) or ATP-bound DnaA (open circle) were measured with a filter binding assay. C, dissociation of ATP-DnaA from other bacteria. B. subtilis DnaA (18 pmol, open circle), S. pyogenes DnaA (18 pmol, closed circle), or E. coli DnaA (18 pmol, open square) was preincubated with 1 μm [α-32P]ATP on ice and then mixed with 1 mm ATP and incubated at 37 °C for indicated time.
Melting of Duplex oriC by S. aureus DnaA
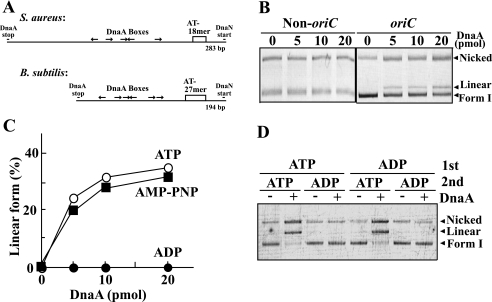
The low nucleotide exchange rate or the stability of the E. coli ADP-DnaA is thought to account for the predominance of ADP-DnaA in E. coli (10, 12). Given the relatively high rate of nucleotide exchange or the instability of the S. aureus ADP-DnaA, whether this protein is actually regulated by adenine nucleotide binding is not clear. Therefore, we examined the effect of nucleotide exchange on the ability of S. aureus DnaA activity to melt the oriC duplex in vitro. Duplex opening at a B. subtilis oriC in vitro occurred at the 27-mer AT-cluster between the DnaA and DnaN ORFs (Fig. 2A) (52, 53). The gap region between dnaA and dnaN of S. aureus has an 18-mer AT-cluster along with five DnaA boxes, which closely resembles the structure and arrangement of the corresponding oriC region of B. subtilis (Fig. 2A) (53, 54). Therefore, to measure open complex formation by S. aureus DnaA, we carried out P1 nuclease assays using pCK206 DNA, which contains this DNA sequence from S. aureus in the pCK20 plasmid. P1 nuclease is a single-stranded DNA-specific endonuclease, which can cleave a single-stranded region induced by DnaA and convert a covalently closed circular form of plasmid (Form I) into linear form (55). The pCK206 DNA was incubated with S. aureus DnaA and HU proteins at 37 °C for 10 min and then digested with P1 nuclease. The linear form of pCK206 DNA was generated in a DnaA-dependent manner (Fig. 2B). A linear form was not generated when a non-oriC plasmid, pCK20, was used for the assay, suggesting that formation of this linear form depends on oriC (Fig. 2B). S. aureus HU protein is a factor that increases the specificity of the cleavage for both oriC and DnaA (data not shown).
FIGURE 2.
Reactivation of the ADP-bound form of S. aureus DnaA in open complex formation in vitro. A, schematic representation of the S. aureus origin region and the corresponding region in B. subtilis. The DnaA box in S. aureus has the consensus sequence 5′-TTATNCACA-3′ or this sequence with a one-base change. The B. subtilis DnaA box was obtained from previous reports (53, 54). B, pCK206 (100 fmol) (oriC) or pCK20 (non-oriC) DNA preincubated with S. aureus HU protein (100 ng) was incubated with S. aureus DnaA at 37 °C for 10 min, digested with P1 nuclease, and analyzed by agarose gel electrophoresis. C, pCK206 DNA (100 fmol) was incubated with S. aureus DnaA in the presence of 5 mm ATP (open circles), 5 mm ADP (closed circles), or 5 mm of the non-hydrolyzable ATP analog AMP-PNP (close squares). Open complex formation was assayed as in B, and the percentage of plasmid in the linear form was calculated from intensities of the plasmid bands determined by agarose gel electrophoresis. D, S. aureus DnaA (15 pmol) was preincubated in a 5-μl solution containing 50 μm ATP or ADP and then added to a 50-μl solution containing pCK206 DNA (100 fmol) and 5 mm ATP or ADP. Open complex formation was assayed as in B.
To examine whether this cleavage took place at the AT-cluster region of oriC, we isolated the plasmid DNA after P1 nuclease digestion and cut it with an endonuclease. Digestion with HindIII or StuI produced 2.7- and 0.7-kbp fragments or 2.1- and 1.3-kbp fragments, respectively (data not shown), which is consistent with P1 nuclease cleavage of pCK206 at or near the AT-cluster region of oriC (data not shown). This suggests that S. aureus DnaA melted the oriC duplex at the 18-mer AT-cluster region in vitro.
Next, we investigated whether the ability of S. aureus DnaA to form an open complex requires ATP and its hydrolysis. In the presence of 5 mm ATP, S. aureus DnaA stimulated formation of the linear form of the pCK206 oriC plasmid, whereas, 5 mm ADP did not (Fig. 2C). Further, AMP-PNP, a non-hydrolyzable analog of ATP, stimulated formation of the linear form of the oriC plasmid by DnaA (Fig. 2C). These results suggest that the open complex formation at oriC by S. aureus DnaA requires ATP but not ATP hydrolysis.
The concentration of ATP required for open complex formation by S. aureus DnaA varied according to the concentration of Mg2+. The standard buffer containing 10 mm Mg2+, DnaA required 1 mm ATP to promote open complex formation, whereas in another buffer containing 0.1 mm Mg2+ and 0.125 mm EDTA, 10 μm ATP was enough for open complex formation (data not shown). Although previous reports showed that open complex formation by E. coli DnaA in the presence of 8 mm Mg2+ requires millimolar concentrations of ATP (6, 51, 55), our results suggest that a decrease in Mg2+ might also reduce the concentration of ATP needed for open complex formation by E. coli DnaA. Together with the notion that pre-priming complex formation by E. coli DnaA, DnaB, and DnaC takes place at 30 μm ATP (6), DnaA seems not require millimolar concentrations of ATP to initiate oriC replication.
Regeneration of the ADP-bound Form of S. aureus DnaA in Vitro
As shown in Fig. 1, bound ADP dissociates rapidly from S. aureus DnaA, and ADP bound to DnaA is rapidly exchanged for ATP in the presence of an excess of ATP. Thus, we investigated whether the rapid exchange of ADP for ATP activates S. aureus DnaA in vitro. As expected, DnaA preincubated with ADP induced melting at oriC when the reaction was done in the buffer with 5 mm ATP (Fig. 2D). In reverse, DnaA preincubated with ATP did not show open complex formation in buffers containing excess ADP (Fig. 2D). The linear form was formed, when the AMP-PNP/total adenine nucleotides ratio was over 30% in open complex formation reactions (data not shown). This was observed at not only 5 mm but also 1 mm total adenine nucleotides (data not shown), thereby the requirement of >30% ATP was not due to necessity of millimolar concentrations of ATP. These results suggest that the rapid exchange of bound ADP with ATP restores the replication initiation activity of DnaA. This is very different from E. coli DnaA, where the ADP-bound form is highly stable and reactivation does not take place easily, even in the presence of excess ATP.
Biochemical Characterization of the R318H Mutant of DnaA
We next tried to construct a mutated DnaA with greatly reduced intrinsic ATPase activity to determine whether DnaA is inactivated by ADP binding in S. aureus. Mutation of Arg-334 of the E. coli AAA+ sensor 2 mutated protein to His or Ala eliminates its intrinsic ATPase activity, and expression of the R334A mutant in E. coli cells causes excessive initiation of chromosome replication (7, 56). We therefore mutated the equivalent residue, Arg-318, of S. aureus DnaA to His. The R318H mutant protein was expressed and purified using the same protocol employed for wild-type S. aureus DnaA (Fig. 3A). Purified DnaAR318H had high affinities for ATP and ADP, with Kd values of 21 and 25 nm, respectively, which are similar to the values for wild-type DnaA (5 nm for ATP and 14 nm for ADP, respectively). The stoichiometry of ATP binding by wild-type and R318H was ∼0.2 and 0.1, respectively, although the value varied between batches. The rate of ATP dissociation from the ATP-DnaAR318H complex at 37 °C was slightly higher than that of wild-type DnaA (Fig. 3B). The intrinsic ATPase activity of R318H was 6% of that of wild-type DnaA, which was determined from their initial velocities (Fig. 3C). Hydrolysis of ATP by wild-type S. aureus DnaA was very slow and was stimulated by plasmid DNA without any apparent specificity for oriC sequence (data not shown), like E. coli DnaA (6). Regardless, like the wild-type form, the R318H mutant DnaA promoted ATP-dependent open complex formation at oriC (Fig. 3D). Therefore, it appears that the R318H protein can initiate DNA replication despite having greatly reduced ATPase activity.
Inhibition of Colony Formation and Excessive Initiation by DnaAR318H in S. aureus Cells
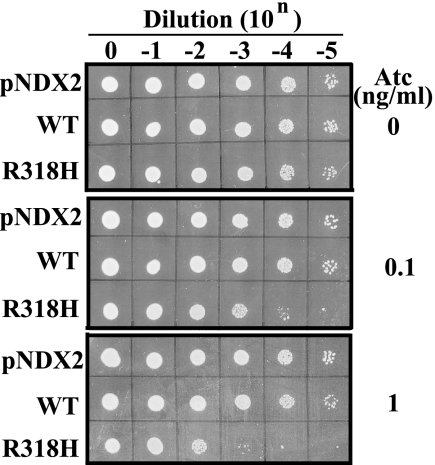
Next, we determined whether DnaAR318H induces excessive initiation of chromosome replication in S. aureus cells. In the case of E. coli, intrinsic ATPase activity-deficient DnaA mutant inhibited the colony formation of E. coli (7). So, we first established cells in which wild-type or R318H DnaA could be conditionally expressed in S. aureus cells by placing the ORFs under control of the Atc-inducible xyl-tet promoter-operator fusion (48). On an LB agar plate containing 0.1 or 1 ng/ml Atc, expression of R318H inhibited the colony formation of the strain, although wild type did not show any effect (Fig. 4). To address the levels of overexpression, we performed Western blotting for total DnaA protein levels from the cells treated with 1 ng/ml Atc (Table 1). DnaA protein levels were increased 1.5- and 1.6-fold (relative to cells with empty vector) in the cells with wild-type versus R318H DnaA-containing plasmids, respectively, suggesting that the inhibition of colony formation by R318H expression was not due to excessive synthesis of the mutant protein.
FIGURE 4.
Inhibition of colony formation by induction of DnaAR318H in S. aureus. An overnight culture of S. aureus NM1003 cells harboring pNDX2-dnaA, pNDX2-R318H, or pNDX2 vector was serially diluted, and 1-μl samples were spotted on an LB agar plate containing 0 (top panel), 0.1 (middle panel), or 1 (bottom panel) ng/ml Atc. The plates were incubated at 37 °C for 20 h and scanned. Results are representative of at least three independent experiments.
TABLE 1.
Relative amounts of DnaA in S. aureus after induction with Atc
| Atca | pNDX2 | WT | R318H |
|---|---|---|---|
| ng/ml | |||
| 0 | 1.0 | 0.8 | 0.6 |
| 1 | 0.9 | 1.5 | 1.6 |
| 10 | 1.0 | 11.0 | 9.4 |
aS. aureus NM1003 cells harboring empty pNDX2 vector, pNDX2-dnaA, or pNDX2-R318H were grown at 37 °C in LB medium until the A600 reached 0.1. Production of DnaA was induced by the addition of Atc and incubation at 37 °C for 1 h. The amount of DnaA after induction was determined by Western blotting and is shown relative to the level in NM1003 cells harboring pNDX2 at 0 ng/ml Atc.
We next measured the effect of expressing R318H DnaA on the origin-to-terminus ratio to know the R318H DnaA expression induce excessive initiation of DNA replication in S. aureus. Southern blotting revealed that induction of R318H by 1 ng/ml Atc increased the origin-to-terminus ratio, whereas expression of the wild-type protein did not (Fig. 5). These results suggest that expression of R318H induces excessive initiation of chromosome replication in S. aureus cells. Therefore, similar to E. coli DnaA, exchange from ATP-DnaA to ADP-DnaA is crucial for negative regulation for the initiation of chromosome replication in S. aureus. On the other hand, a little increase in origin-to-terminus ratio after induction of wild-type S. aureus DnaA differs from similar studies with E. coli DnaA (7, 57), which might be due to differences in initiation control of chromosome replication between them.
FIGURE 5.
Excessive initiation in vivo by S. aureus DnaAR318H. S. aureus NM1003 harboring pNDX2-dnaA (closed circles) or pNDX2-R318H (open circles) was grown until the A600 reached 0.1, adjusted to 1 ng/ml Atc, and incubated at 37 °C for the indicated time. The ratio of replication origin to terminus was determined by Southern hybridization. Results are means ± S.E. of at least three independent experiments.
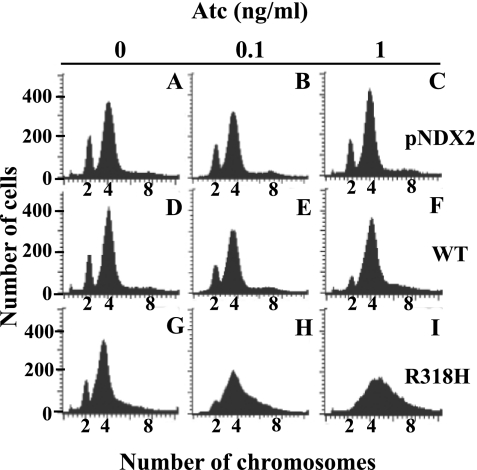
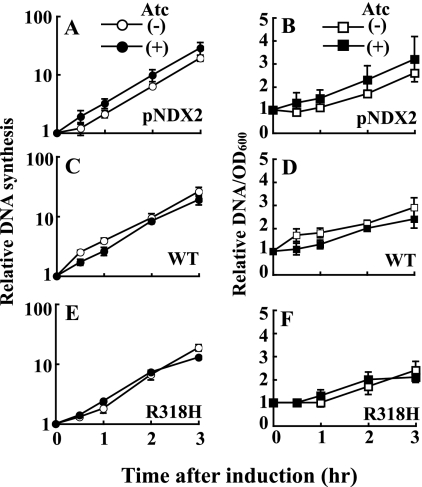
In addition, using flow cytometry, we measured the effect of R318H DnaA expression on the chromosome equivalents (58). Exponentially growing cells were treated with 0.1 or 1 ng/ml of Atc for 1 h, followed by 2 h with rifampicin and cephalexin, which allows completion of ongoing rounds of chromosome replication without a new round of initiation and cell division. Cells harboring the wild-type DnaA-expressing plasmid or an empty vector showed major 4N and minor 2N peaks in the absence or presence of 0.1 or 1 ng/ml of Atc (Fig. 6, A–F). These peaks were not formed in cells harboring the R318H treated with 1 ng/ml of Atc; instead, a broaden peak was observed, in which most of the cells contained chromosome equivalents of between 4N and 8N (Fig. 6I), whereas cells harboring the R318H without Atc showed major 4N and minor 2N peaks (Fig. 6G). This suggests that ongoing rounds of replication are not completed in the R318H-expressing cells. Consistent with this notion, the relative uptake of [3H]thymine into acid-insoluble materials (relative to turbidity) after induction of R318H expression by 1 ng/ml Atc (Fig. 7F) was not higher than that in control strains, but was rather similar to that in control strains (Fig. 7, B or D). Therefore, it appears that R318H expression induces excessive initiation from oriC but results in incompletion of the following replication of entire chromosome, in which only oriC and its neighboring region may be replicated as suggested after induction of excessive initiation by E. coli DnaAA184V and DnaAR334A mutants (7, 12, 59).
FIGURE 6.
Expression of DnaAR318H prevents replication fork movement in vivo. S. aureus NM1003 cells harboring empty pNDX2 vector (A–C), pNDX2-dnaA (D–F), or pNDX2-R318H (G–I) were grown until the A600 reached at 0.1, mixed with 0 (A, D, and G), 0.1 (B, E, and H), or 1 (C, F, and I) ng/ml Atc and incubated at 37 °C for 1 h. Cells were further incubated for 2 h in the presence of rifampicin and cephalexin and then harvested. Chromosome equivalents of cells were analyzed by flow cytometry using a FACSCalibur.
FIGURE 7.
Chromosomal DNA replication in DnaAR318H-expressing cells. S. aureus NM1003 cells harboring empty pNDX2 vector (A and B), pNDX2-dnaA (C and D), or pNDX2-R318H (E and F) were grown at 37 °C in LB medium containing [3H]thymine until the A600 reached 0.2. One half of the culture was supplemented with 1 ng/ml Atc (closed symbols), and the other half was not treated with Atc (open symbols). The incubation was then continued at 37 °C, and, at the indicated times, portions were withdrawn for measurement of the optical density and the incorporation of [3H]thymine into acid-insoluble materials (left panels). Relative amounts of [3H]thymine incorporation to A600 value are represented (right panels). Results are means ± S.E. of at least three independent experiments.
When we carefully compared Fig. 6D with Fig. 6F, we could find a slightly higher 4N-to-2N ratio in Fig. 6F, suggesting induction of wild-type DnaA caused more frequent initiations. Although this observation appears to contradict the results in Fig. 5, it may be explained by different sensitivities of the two measurements, Southern blotting evaluating the average number in Fig. 5 and flow cytometry evaluating single cell in a highly sensitive manner in Fig. 6. An average origin number per cell calculated from Fig. 6 is 3.6 and 3.9 in Fig. 6D and Fig. 6F, respectively, demonstrating that the average increase in origin number per cell by the DnaA induction is <10%, which is a small change in the measurement of origin-to-terminus ratio in Fig. 5.
Effect of Multicopy Supply of DnaN on R318H-mediated Inhibition of Colony Formation
In B. subtilis, overexpression of wild-type DnaA inhibits expression of dnaN, which induces the SOS response and inhibits cell growth (24). Therefore, we determined whether overexpression of DnaN could suppress the inhibition of colony formation caused by S. aureus DnaAR318H. For these experiments, we constructed a multicopy plasmid for expressing dnaN under control of the hisS promoter. The hisS promoter does not contain a DnaA box (either consensus or a one-base-altered) and therefore is not affected by the amount of DnaA. The resulting plasmid, pSE-dnaN, suppressed temperature-sensitive colony formation by an S. aureus dnaN mutant (data not shown), which we had previously isolated during our S. aureus temperature-sensitive mutant screening project (36, 37, 44), suggesting that pSE-dnaN mediated the expression of sufficient amounts of functional DnaN protein for the growth or viability of the mutant cells. We introduced pSE-dnaN into R318H-expressing S. aureus cells. The inhibition of colony formation and the incompletion of ongoing rounds of replication as detected by flow cytometry caused by induction R318H protein with 0.1 or 1 ng/ml Atc were not suppressed by pSE-dnaN (data not shown). Therefore, it appears that the inhibition of colony formation and the incompletion of ongoing rounds of replication by R318H in S. aureus are not due to insufficient expression of DnaN.
DISCUSSION
In the present study, we investigated the role of the adenine nucleotide binding of S. aureus DnaA in the control of the initiation step of chromosome replication. In vitro, S. aureus DnaA promoted open complex formation at oriC in the presence of ATP but not in the presence of ADP. An R318H AAA+ sensor 2-mutated protein, which had greatly reduced ATPase activity, promoted ATP-dependent open complex formation in vitro and induced excessive initiation of chromosome replication in vivo. Therefore, DnaA activity is negatively regulated by ADP binding, and the ratio of the ATP-bound to ADP-bound form of DnaA may control the initiation step of chromosome replication in S. aureus cells as it does in E. coli DnaA.
Despite these similarities, the present study also revealed some different features of E. coli and S. aureus DnaA. Specifically, the adenine nucleotide bound to S. aureus DnaA was unstable, although the affinities to adenine nucleotides to S. aureus DnaA are almost the same as E. coli DnaA. Due to this instability, ADP-DnaA immediately restored the melting ability at oriC in conditions containing excess ATP than ADP. Because the ATP concentration in cells is much higher than that of ADP, ADP bound to DnaA may be rapidly exchanged for ATP in vivo. This would preclude control of replication initiation by nucleotide binding unless there are some regulatory systems for controlling nucleotide binding by DnaA. In other words, S. aureus should have some mechanisms for increasing the intracellular concentration of ADP-DnaA to suppress replication initiation. One possible mechanism is the stimulation of the intrinsic ATPase activity of DnaA, which would be similar to the mechanism of RIDA in E. coli (10, 13). Alternatively, there could be a factor that inhibits dissociation of bound adenine nucleotides from DnaA, similar to factors that inhibit the release of guanine nucleotides by small G proteins (60, 61). Conversely, S. aureus may use this biochemical property of DnaA, immediate rejuvenation, for the timely activation of DnaA to initiate chromosome replication in cells.
We estimated that each S. aureus cell contains 700 ± 100 DnaA molecules (44), and 50–100 DnaA molecules are needed to melt one duplex of oriC (Fig. 2). Therefore, it appears that most of the DnaA molecules in an S. aureus cell are in an inactive state to prevent excessive initiation (62). This agrees with the conclusion that S. aureus DnaA is inactivated by generation of the ADP-bound form.
Recently, extensive studies found that replication initiators in eubacteria, archea, and eukaryotes depend on ATP. In eukaryotes, initiation of replication by the yeast origin recognition complex (Orc) depends on ATP hydrolysis (63, 64). The ADP-bound form of Orc1p is thought to dissociate the initiation complex and promote the replication process by a mechanical process rather than acting as a suppressor replication initiation (65). Because the ATPase-deficient R318H mutant of S. aureus DnaA caused excessive replication initiation, like E. coli DnaA, it appears to act as a regulator of replication initiation rather than by mechanically influencing replication. Together, these findings suggest that ATP hydrolysis by initiator proteins plays different roles in prokaryote and eukaryotes.
Acknowledgments
We thank Drs. W. Hillen, S. Yasuda, N. Ogasawara, and J. J. Ferretti for kindly providing pWH353 and pWH354, pBAD24, pMUTIN-T3, and pSF151, respectively. We are grateful to Drs. S. Kawabata and H. Yoshikawa for bacteria. We also thank Dr. C. Kaito for his construction of pCK206, Dr. M. Matsuo for her construction of pKE516 and isolation of an S. aureus dnaN mutant, and M. Miyatani and L. Reutimann for their technical assistance.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science, by the Industrial Technology Research Grant Program of the NEDO of Japan (2004), and by grants from Kyorin Pharmaceutical Co., Ltd. and Genome Pharmaceuticals Co., Ltd.
M. Matsuo, K. Kurokawa, and K. Sekimizu, unpublished observations.
- RIDA
- regulatory inactivation of DnaA
- Atc
- anhydrotetracycline
- ORF
- open reading frame
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- Orc
- origin recognition complex
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
REFERENCES
- 1.Skarstad K., Boye E. (1994) Biochim. Biophys. Acta 1217, 111–130 [DOI] [PubMed] [Google Scholar]
- 2.Donachie W. D., Blakely G. W. (2003) Curr. Opin Microbiol. 6, 146–150 [DOI] [PubMed] [Google Scholar]
- 3.Messer W. (2002) FEMS Microbiol. Rev. 26, 355–374 [DOI] [PubMed] [Google Scholar]
- 4.Kornberg A., Baker T. (1992) DNA Replication, Second Ed., W. H. Freeman and Company, New York [Google Scholar]
- 5.Kaguni J. M. (2006) Annu. Rev. Microbiol. 60, 351–371 [DOI] [PubMed] [Google Scholar]
- 6.Sekimizu K., Bramhill D., Kornberg A. (1987) Cell 50, 259–265 [DOI] [PubMed] [Google Scholar]
- 7.Nishida S., Fujimitsu K., Sekimizu K., Ohmura T., Ueda T., Katayama T. (2002) J. Biol. Chem. 277, 14986–14995 [DOI] [PubMed] [Google Scholar]
- 8.Erzberger J. P., Mott M. L., Berger J. M. (2006) Nat. Struct. Mol. Biol. 13, 676–683 [DOI] [PubMed] [Google Scholar]
- 9.Mott M. L., Erzberger J. P., Coons M. M., Berger J. M. (2008) Cell 135, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999) EMBO J. 18, 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S. Y., Yokoyama S., Kurumizaka H., Katayama T. (2008) J. Biol. Chem. 283, 8351–8362 [DOI] [PubMed] [Google Scholar]
- 12.Katayama T. (2001) Mol. Microbiol. 41, 9–17 [DOI] [PubMed] [Google Scholar]
- 13.Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. (1998) Cell 94, 61–71 [DOI] [PubMed] [Google Scholar]
- 14.Kato J., Katayama T. (2001) EMBO J. 20, 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su'etsugu M., Shimuta T., Ishida T., Kawakami H., Katayama T. (2005) J. Biol. Chem. 280, 6528–6536 [DOI] [PubMed] [Google Scholar]
- 16.Camara J. E., Breier A. M., Brendler T., Austin S., Cozzarelli N. R., Crooke E. (2005) EMBO Rep. 6, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riber L., Olsson J. A., Jensen R. B., Skovgaard O., Dasgupta S., Marinus M. G., Løbner-Olesen A. (2006) Genes Dev. 20, 2121–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su'etsugu M., Nakamura K., Keyamura K., Kudo Y., Katayama T. (2008) J. Biol. Chem. 283, 36118–36131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekimizu K., Kornberg A. (1988) J. Biol. Chem. 263, 7131–7135 [PubMed] [Google Scholar]
- 20.Zheng W., Li Z., Skarstad K., Crooke E. (2001) EMBO J. 20, 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima T., Ishikawa Y., Obana E., Hase M., Kubota T., Katayama T., Kunitake T., Watanabe E., Sekimizu K. (1996) J. Biol. Chem. 271, 3633–3638 [DOI] [PubMed] [Google Scholar]
- 22.Fujimitsu K., Senriuchi T., Katayama T. (2009) Genes Dev. 23, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun R. E., O'Day K., Wright A. (1985) Cell 40, 159–169 [DOI] [PubMed] [Google Scholar]
- 24.Ogura Y., Imai Y., Ogasawara N., Moriya S. (2001) J. Bacteriol. 183, 3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa S., Ogura Y., Yoshimura M., Okumura H., Cho E., Kawai Y., Kurokawa K., Oshima T., Ogasawara N. (2007) DNA Res. 14, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goranov A. I., Katz L., Breier A. M., Burge C. B., Grossman A. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gon S., Camara J. E., Klungsøyr H. K., Crooke E., Skarstad K., Beckwith J. (2006) EMBO J. 25, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speck C., Weigel C., Messer W. (1999) EMBO J. 18, 6169–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M., Campbell J. L., Boye E., Kleckner N. (1994) Cell 77, 413–426 [DOI] [PubMed] [Google Scholar]
- 30.Slater S., Wold S., Lu M., Boye E., Skarstad K., Kleckner N. (1995) Cell 82, 927–936 [DOI] [PubMed] [Google Scholar]
- 31.Brendler T., Abeles A., Austin S. (1995) EMBO J. 14, 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriya S., Imai Y., Hassan A. K. M., Ogasawara N. (1999) Plasmid 41, 17–29 [DOI] [PubMed] [Google Scholar]
- 33.Velten M., McGovern S., Marsin S., Ehrlich S. D., Noirot P., Polard P. (2003) Mol. Cell 11, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 34.Rokop M. E., Auchtung J. M., Grossman A. D. (2004) Mol. Microbiol. 52, 1757–1767 [DOI] [PubMed] [Google Scholar]
- 35.Marsin S., McGovern S., Ehrlich S. D., Bruand C., Polard P. (2001) J. Biol. Chem. 276, 45818–45825 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Kurokawa K., Reutimann L., Mizumura H., Matsuo M., Sekimizu K. (2007) Microbiology 153, 3370–3379 [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Kurokawa K., Matsuo M., Fukuhara N., Murakami K., Sekimizu K. (2004) Mol. Genet. Genomics 271, 447–457 [DOI] [PubMed] [Google Scholar]
- 38.Dervyn E., Suski C., Daniel R., Bruand C., Chapuis J., Errington J., Jannière L., Ehrlich S. D. (2001) Science 294, 1716–1719 [DOI] [PubMed] [Google Scholar]
- 39.Inoue R., Kaito C., Tanabe M., Kamura K., Akimitsu N., Sekimizu K. (2001) Mol. Genet. Genomics 266, 564–571 [DOI] [PubMed] [Google Scholar]
- 40.Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T. (2007) Genes Dev. 21, 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noirot-Gros M. F., Dervyn E., Wu L. J., Mervelet P., Errington J., Ehrlich S. D., Noirot P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noirot-Gros M. F., Velten M., Yoshimura M., McGovern S., Morimoto T., Ehrlich S. D., Ogasawara N., Polard P., Noirot P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Defeu Soufo C., Defeu Soufo H. J., Noirot-Gros M. F., Steindorf A., Noirot P., Graumann P. L. (2008) Dev. Cell 15, 935–941 [DOI] [PubMed] [Google Scholar]
- 44.Murai N., Kurokawa K., Ichihashi N., Matsuo M., Sekimizu K. (2006) FEMS Microbiol. Lett. 254, 19–26 [DOI] [PubMed] [Google Scholar]
- 45.Yamagishi J., Kojima T., Oyamada Y., Fujimoto K., Hattori H., Nakamura S., Inoue M. (1996) Antimicrob. Agents Chemother. 40, 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichihashi N., Kurokawa K., Matsuo M., Kaito C., Sekimizu K. (2003) J. Biol. Chem. 278, 28778–28786 [DOI] [PubMed] [Google Scholar]
- 47.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geissendörfer M., Hillen W. (1990) Appl. Microbiol. Biotechnol. 33, 657–663 [DOI] [PubMed] [Google Scholar]
- 49.Ogata Y., Inoue R., Mizushima T., Kano Y., Miki T., Sekimizu K. (1997) Biochim. Biophys. Acta 1353, 298–306 [DOI] [PubMed] [Google Scholar]
- 50.Broyles S. S., Pettijohn D. E. (1986) J. Mol. Biol. 187, 47–60 [DOI] [PubMed] [Google Scholar]
- 51.Sekimizu K., Bramhill D., Kornberg A. (1988) J. Biol. Chem. 263, 7124–7130 [PubMed] [Google Scholar]
- 52.Moriya S., Firshein W., Yoshikawa H., Ogasawara N. (1994) Mol. Microbiol. 12, 469–478 [DOI] [PubMed] [Google Scholar]
- 53.Krause M., Rückert B., Lurz R., Messer W. (1997) J. Mol. Biol. 274, 365–380 [DOI] [PubMed] [Google Scholar]
- 54.Fukuoka T., Moriya S., Yoshikawa H., Ogasawara N. (1990) J. Biochem. 107, 732–739 [DOI] [PubMed] [Google Scholar]
- 55.Bramhill D., Kornberg A. (1988) Cell 52, 743–755 [DOI] [PubMed] [Google Scholar]
- 56.Su'etsugu M., Kawakami H., Kurokawa K., Kubota T., Takata M., Katayama T. (2001) Mol. Microbiol. 40, 376–386 [DOI] [PubMed] [Google Scholar]
- 57.Løbner-Olesen A., Skarstad K., Hansen F. G., von Meyenburg K., Boye E. (1989) Cell 57, 881–889 [DOI] [PubMed] [Google Scholar]
- 58.Skarstad K., Bernander R., Boye E. (1995) Methods Enzymol. 262, 604–613 [DOI] [PubMed] [Google Scholar]
- 59.Simmons L. A., Breier A. M., Cozzarelli N. R., Kaguni J. M. (2004) Mol. Microbiol. 51, 349–358 [DOI] [PubMed] [Google Scholar]
- 60.Bourne H. R., Sanders D. A., McCormick F. (1991) Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 61.Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 62.Mahaffy J. M., Zyskind J. W. (1989) J. Theor. Biol. 140, 453–477 [DOI] [PubMed] [Google Scholar]
- 63.Bowers J. L., Randell J. C. W., Chen S., Bell S. P. (2004) Mol. Cell 16, 967–978 [DOI] [PubMed] [Google Scholar]
- 64.Randell J. C. W., Bowers J. L., Rodríguez H. K., Bell S. P. (2006) Mol. Cell 21, 29–39 [DOI] [PubMed] [Google Scholar]
- 65.Takenaka H., Makise M., Kuwae W., Takahashi N., Tsuchiya T., Mizushima T. (2004) J. Mol. Biol. 340, 29–37 [DOI] [PubMed] [Google Scholar]