Abstract
Gap junctions are intercellular channels that allow the passage of ions, small molecules, and second messengers that are essential for the coordination of cellular function. They are formed by two hemichannels, each constituted by the oligomerization of six connexins (Cx). Among the 21 different human Cx isoforms, studies have suggested that in the heart, Cx40 and Cx43 can oligomerize to form heteromeric hemichannels. The mechanism of heteromeric channel regulation has not been clearly defined. Tissue ischemia leads to intracellular acidification and closure of Cx43 and Cx40 homomeric channels. However, coexpression of Cx40 and Cx43 in Xenopus oocytes enhances the pH sensitivity of the channel. This phenomenon requires the carboxyl-terminal (CT) part of both connexins. In this study we used different biophysical methods to determine the structure of the Cx40CT and characterize the Cx40CT/Cx43CT interaction. Our results revealed that the Cx40CT is an intrinsically disordered protein similar to the Cx43CT and that the Cx40CT and Cx43CT can interact. Additionally, we have identified an interaction between the Cx40CT and the cytoplasmic loop of Cx40 as well as between the Cx40CT and the cytoplasmic loop of Cx43 (and vice versa). Our studies support the “particle-receptor” model for pH gating of Cx40 and Cx43 gap junction channels and suggest that interactions between cytoplasmic regulatory domains (both homo- and hetero-connexin) could be important for the regulation of heteromeric channels.
Introduction
The synchronization of myocardial contraction and propagation of the cardiac action potential require proper intercellular communication (1, 2). This intercellular communication can be altered, for example, by an inadequate blood flow from constriction or blockage of blood vessels supplying the heart, leading the cardiac tissue to become hypoxic or in the worst case, anoxic. This metabolic abnormality called ischemia induces intracellular acidification and exerts effects on cardiac contractility and rhythm. These arrhythmias are caused in part by alterations in the expression, localization, and regulation of the gap junctions proteins, which are responsible for the intercellular communication (3, 4). Although gap junctions can be regulated in a variety of ways (e.g. Ca+2, pH, and phosphorylation state), intracellular acidification initially leads to closure of gap junctions (5–7) before any detectable changes in the integrity of the gap junction plaque (8), suggesting that gap junction channels close while still at the membrane and before being internalized (9). Acidification-induced uncoupling also has an impact on the preservation of tissue surrounding the ischemic area in the heart and may be a key substrate for life-threatening arrhythmias during myocardial infarction (10).
Gap junctions are formed by the apposition of connexons from adjacent cells, where each connexon (or hemi-channel) is formed by six connexin proteins. Connexins are tetraspan transmembrane domain proteins with intracellular amino and carboxyl termini. There are 21 different connexin genes in the human genome. The major divergence in primary structures, which allow specific regulatory properties for each isoform, occurs in the cytoplasmic loop (CL)3 and carboxyl-terminal (CT) domains. Each of the connexins can form gap junctions by themselves; however, studies have demonstrated that many cells co-express more than one connexin isoform, giving rise to heteromeric connexons (i.e. more than one connexin isotype in the same single connexon; for review, see Ref. 11).
Connexin43 (Cx43) and connexin40 (Cx40) co-express in several tissues, including cardiac atrial and ventricular myocytes, and vascular smooth muscle (12). The intercellular communication provided by Cx43 and Cx40 gap junctions is a principal determinant of myocardial conduction, and altered expression has been implicated in arrhythmogenesis (13). Cx43 and Cx40 can hetero-oligomerize, leading to the formation of heterologous gap junctions (12, 14–16). The physiological consequences of hetero-oligomerization on gap junction intercellular communication remain unclear, although functional studies demonstrate that these structures display biophysical properties that do not correspond to their homomeric forms (14, 15, 17–20). Because the sensitivity of different homomeric gap junctions to biochemical modulators varies widely, the question arises as to what types of interactions occur between connexins to integrate regulatory signals in heteromeric channels. Evidence supports involvement of the CT domains in regulation of these heteromeric channels.
Junctional conductance studies between Xenopus oocyte pairs have shown that pH regulation and observation of the lower conductance state of Cx43 and Cx40 depend on the presence of the CT domain (7, 15, 21, 22). Replacing Cx40CT with the CT domain of Cx43 rescues the behavior of the homologous channel both when the fragments (i.e. the Cx40 truncated channel and the Cx43CT domain) are separately co-expressed and when both domains are covalently attached (22). Additionally, co-expression of Cx43 and Cx40 in the same oocyte was more susceptible to acidification-induced uncoupling than those cells expressing only one connexin isoform (15). These data support that both Cx43 and Cx40 1) follow the particle-receptor hypothesis for pH gating, 2) interact within a heteromeric connexon in a synergistic manner, and 3) have regulatory domains that can specifically interact with a channel formed by another connexin. These observations combined with our previous studies identifying that Cx43CT dimerization and the Cx43CT/Cx43CL interaction may be some of the structural changes involved in the pH regulation of Cx43 (23–25) have led us to consider that Cx40CT dimerization, Cx40CT/Cx43CT oligomerization, and the Cx40CT/Cx43CL and Cx43CT/Cx40CL interactions are structures involved in pH regulation of homomeric and heteromeric channels involving Cx40.
Here, we have characterized the structure and biophysical properties of the Cx40CT domain and assessed its ability to associate with itself and the Cx43CT as well as the Cx40CL and Cx43CL domains. Nuclear magnetic resonance (NMR) studies determined that the Cx40CT is an intrinsically disordered protein; however, Fourier transform infrared (FTIR) spectroscopy and circular dichroism (CD) suggested that some residues may have a propensity to form a very dynamic α-helix. NMR titration studies in the presence of trifluoroethanol (TFE) identified one Cx40CT region (residues Cys-267—Gly-285) that can be induced to form α-helical structure. Sedimentation equilibrium studies determined that the Cx40CT exists predominately in a monomeric state with a minor population in a dimeric conformation. Furthermore, we have determined that the Cx40CT and Cx43CT, Cx40CT and Cx43CL, Cx43CT and Cx40CL, Cx43CT and Cx43CL, and Cx40CT and Cx40CL domains can associate and have identified the residues involved in these interactions. Our studies support the particle-receptor model for pH gating of Cx40 and Cx43 gap junction channels, and we hypothesize that intermolecular interactions between CT and CL domains occur in functional Cx43/Cx40 channels, which may play a role in the higher pH sensitivity of those heteromeric channels.
MATERIALS AND METHODS
Expression and Purification of Recombinant Glutathione S- Transferase (GST)-Cx40CT and GST-Cx43CT
The Cx40CT (residues 251–355), Cx43CT (residues 255–382), Cx40CL (residues 100–141), and Cx43CL (residues 100–155) polypeptides (unlabeled, 15N-labeled, or 13C15N-labeled) were expressed and purified as described previously (23, 25). All polypeptides were confirmed for purity and analyzed for degradation by SDS-PAGE and mass spectroscopy. All polypeptides were equilibrated in 1× phosphate-buffered saline (PBS) at pH 6.0 and 1 mm dithiothreitol using NAP-5 columns (GE Healthcare).
NMR
All NMR data were acquired at 7 °C using a 600-MHz Varian INOVA NMR spectrometer outfitted with a cryo-probe. Experimental data to determine the Cx40CT and Cx43CL backbone sequential assignments have been described (26). Distance constraints used to determine the Cx40CT structure were derived from nuclear Overhauser effects (NOEs) observed in the 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra, with mixing times of 125 and 150 ms, respectively. NMR spectra were processed using NMRPipe (27) and analyzed using NMRView (28).
Gradient-enhanced two-dimensional 15N-HSQC experiments (29) were used to observe all backbone amide resonances from the 15N-labeled Cx40CT, Cx43CT, and Cx43CL samples in the absence or presence of different combinations of unlabeled Cx43 and Cx40 CT and CL domains. Data were acquired with 1024 complex points in the direct dimension and 512 complex points in the indirect dimension.
Hydrogen bonds were identified on the basis of temperature dependence studies, which follow the amide proton chemical shifts from 7 to 32 °C (30). Any amide proton that shifted ≤5.0 ppb/K was considered likely to be hydrogen-bonded.
Structure Calculation
Model structures were calculated by simulated annealing using torsion angle dynamics as implemented in the program Crystallography and NMR System (31). NOE cross-peaks classified as strong, medium, and weak were converted into distance restraints of 1.8–2.5, 1.8–3.5, and 1.8–5.5 Å, respectively. 3JHNα coupling constants were measured from a three-dimensional HNHA experiment (32) and used directly as constraints. The 10 best energy-minimized structures were evaluated using AQUA and PROCHECK-NMR (33). The coordinates of the Cx40CT domain have been deposited in the Protein Data Bank (2k7m).
FTIR
Cx40CT was prepared at a concentration of 0.7 mm in PBS (pH 6.0) buffer where the H2O was replaced by D2O. The FTIR spectrum was measured in D2O to avoid the strong water band that overlaps with the amide I bands. A total of 60 μl of solution was inserted between CaF2 windows using a 100-μm spacer. Spectra were recorded on a JASCO-610 spectrometer at 20 °C with 1000 interferograms and a resolution of 2 cm−1. The second derivatives of the spectra were calculated with the software provided by JASCO.
CD
CD experiments were performed on a JASCO-810 spectrometer at 7 °C with a 1-mm path length quartz cell using a bandwidth of 4 nm, an integration time of 1 s, and a scan rate of 1.6 nm/s. Each spectrum is the average of 5 scans. All spectra were corrected by subtracting the solvent spectrum acquired under identical conditions. The protein concentration for each sample was 200 μm. All CD data were processed using CDtool (34) and analyzed by Dichroweb (35).
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed using a Beckman Optima XL-I analytical ultracentrifuge and an AN-60Ti rotor. The Cx40CT was analyzed at 20 °C in PBS buffer (pH 6.0 and 7.4) and 1 mm dithiothreitol. Data were collected at three concentrations (A280 = 0.2, 0.4, and 0.8) and three rotor speeds (24,000, 25,000, and 26,000 rpm). Absorbance scans at 280 nm were taken after 22 and 24 h at each speed; it was assumed that equilibrium was reached if the scans were unchanged. Analysis of the sedimentation equilibrium data were performed using the Beckman XL-A/XL-I software package within Microcal, ORIGIN Version 4. Buffer densities were determined using a Mettler DE40 density meter operated at the experimental temperature, and data were analyzed with the program Sednterp Version 1.03. Partial specific volume was calculated in Sednterp from the amino acid composition.
Surface Plasmon Resonance (SPR)
All SPR experiments were performed using a Biacore 3000 at the University of Nebraska Medical Center's Molecular Interaction Facility. Cx43CT in PBS buffer (pH 6.0) was immobilized covalently on the carboxymethyl dextran matrix sensor chip CM-5 using the Thiol Coupling Kit provided by Biacore. Three cysteine residues are naturally present in the Cx43CT sequence. In each flow cell, 2 nmol of protein were loaded onto the surface, resulting in ∼500 “response units” of protein captured.
All analysis were performed at a flow rate of 20 μl/min. Before the loading of analytes, the chip was equilibrated with PBS (pH 6.0) containing 0.005% surfactant P-20 (PBS-P). Each analyte at various concentrations in the same buffer was injected in duplicate at random over the immobilized ligand. After injection of the analyte, PBS-P was reintroduced with a 300-s lag time to start dissociation. The chip was regenerated to base line by injection of 5 μl of 100 mm HCl or 0.1 m NaHCO3 (pH 8.6).
In Vitro GST Pulldown Assay
The ProFound Pulldown GST Protein:Protein Interaction kit (Thermo Scientific) was used to test for the Cx43CT/Cx40CT interaction. Approximately 150 μg of purified GST-Cx43CT or GST-Cx40CT were added to immobilized glutathione and incubated at 4 °C for 30 min. The beads were washed 5× with TBS:ProFound lysis buffer containing 25 mm Tris·HCl, 0.15 m NaCl (pH 7.2) and then incubated with 150 μg of Cx40CT or Cx43CT, respectively, at 4 °C for 1 h. The beads were then washed 5× with TBS:ProFound lysis buffer, and the proteins were eluted with 100 mm glutathione. The proteins were resolved on 10% Tris-Tricine SDS-acrylamide gels and analyzed with either an anti-Cx40CT (Alpha Diagnostic) or anti-Cx43CT (Fred Hutchinson Cancer Research Center) antibody.
RESULTS
Structure Calculation and Description of the Cx40CT
Our laboratory recently published resonance assignments for the Cx40CT (26). We have now applied NOE spectroscopy to identify the resonance peaks that presented stable secondary structures. The solution structure was obtained in PBS (pH 6.0) at 7 °C. Structure calculations by torsion angle dynamics followed with refinement by simulated annealing, and energy minimization led to the family of structures shown in Fig. 1. The overall structure of the Cx40CT (Ser-251–Val-355), as calculated from using 1123 NOE restraints, is intrinsically disordered or random-coiled. Structural statistics are presented in supplemental Table 1. The intrinsically disordered structure for the Cx40CT is consistent with the narrow chemical shift dispersions in the 15N-HSQC (26).
FIGURE 1.
Solution structure of the Cx40CT (Cx40 residues Ser-251–Val-355) in PBS buffer (pH 6.0) at 7 °C. A, shown is the lowest energy structure. B, shown are backbone traces of the 10 Cx40CT NMR conformers that best represent the structure aligned by superimposing the backbone atoms.
Interestingly, the chemical shifts from the NMR temperature coefficient experiment suggest the possibility that Cx40CT residues Asp-279—Ser-286 are involved in hydrogen bonding (supplemental Fig. 1). However, this region did not display the proportion of medium range NOEs (e.g. for an α-helix, (i,i+3) and (i,i+4) NOE connectivities) that would be consistent with stable secondary structure, and this region displayed little to no 13Cα and 13Cβ chemical shift deviations from random coil (supplemental Fig. 2). These hydrogen bond restraints were not included in the modeling because the NOE restraints alone did not model this region as α-helical or β-strands in structure.
Propensity of the Cx40CT to Form α-Helical Structure
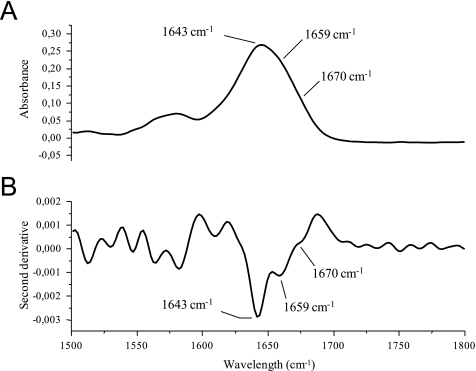
Although no stable secondary structure was observed for the Cx40CT, the observation from the temperature coefficient experiment that Cx40CT residues Asp-279—Ser-286 may form hydrogen bonds suggests that these residues may be rapidly interconverting between unfolded (predominately) and folded (small population) states. Therefore, to identify if a very small amount of a dynamic α-helix is present in the Cx40CT domain, we used FTIR (Fig. 2). We focused on the amide I band (between 1600 and 1700 cm−1), which is formed by C O stretching vibration (70–85%) and C-N groups (10–20%), because the C
O stretching vibration (70–85%) and C-N groups (10–20%), because the C O bond is involved in the hydrogen bonding that takes place between the different elements of secondary structure and, therefore, is sensitive to the secondary structure content of a protein. One common difficulty with analyzing the amide I band for secondary structure is that shifts are small compared with the intrinsic width of the band (Fig. 2A). Therefore, to distinguish the various components of the amide I band, we use the second derivative spectrum (Fig. 2B). A major band appears at 1643 cm−1, which is consistent with random coil structure. A weaker absorption at 1659 cm−1 may correspond to a very dynamic α-helix accessible to the solvent. Also, a very broad band at 1670 cm−1 corresponds to turns typically present in dynamic and unfolded proteins.
O bond is involved in the hydrogen bonding that takes place between the different elements of secondary structure and, therefore, is sensitive to the secondary structure content of a protein. One common difficulty with analyzing the amide I band for secondary structure is that shifts are small compared with the intrinsic width of the band (Fig. 2A). Therefore, to distinguish the various components of the amide I band, we use the second derivative spectrum (Fig. 2B). A major band appears at 1643 cm−1, which is consistent with random coil structure. A weaker absorption at 1659 cm−1 may correspond to a very dynamic α-helix accessible to the solvent. Also, a very broad band at 1670 cm−1 corresponds to turns typically present in dynamic and unfolded proteins.
FIGURE 2.
FTIR of the Cx40CT demonstrating a small amount of a dynamic α-helix. A, shown is a FTIR absorption spectrum of the Cx40CT in D2O PBS. B, shown is the second-derivative spectrum of panel A. Bands corresponding to the random coil structure (strong, 1643 cm−1) and dynamic α-helical structure (weak, 1659 cm−1) and turns typically present in dynamic and unfolded proteins (very weak, 1670 cm−1) have been labeled.
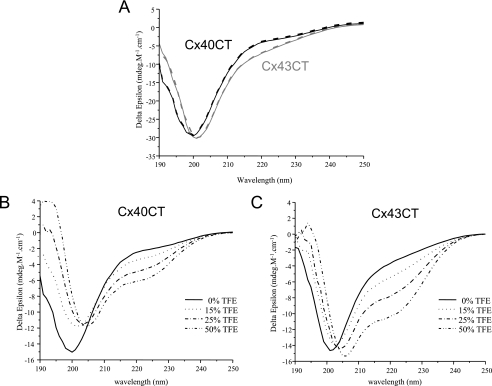
To complement the FTIR experiment, we used CD spectroscopy in the far-UV spectral region (190–250 nm) to analyze the Cx40CT peptide bonds. Additionally, as a comparison we also performed CD on the Cx43CT domain, where previous structural studies identify two small α-helical domains in an otherwise predominately random-coiled structure (24). As expected from the NMR data, Cx40CT and Cx43CT at pH 6.0 present a high proportion of random-coiled structure (major absorption at 200 nm for Cx40CT and 201 nm for Cx43CT) (Fig. 3A). An absorption at 222 nm appears for Cx43CT, corresponding to the specific absorption of α-helices in agreement with the NMR structure (36). This observation is also consistent with the small shift observed between the maximum absorption at 200 nm for Cx40CT to 201 nm for Cx43CT, probably due to the characteristic contribution of α-helix signal at 208 nm. Although the absorption intensity at 222 nm indicates that the Cx43CT contains more α-helical content than Cx40CT, a weak but measurable absorption at 222 nm for Cx40CT is evident. This suggests, similar to the FTIR data, the possibility of a small amount of a very dynamic α-helix. Additionally, pH has no effect on the Cx43CT or Cx40CT secondary structure content (data not shown).
FIGURE 3.
CD spectra of Cx40CT and Cx43CT. A, CD spectra of Cx40CT (in black) and Cx43CT (in gray) were performed in PBS buffer at pH 6.0. CD spectra of Cx40CT (B) and Cx43CT (C) were obtained in the presence of varying concentrations of TFE (% displayed in each panel).
TFE is a well known α-helix stabilizer; therefore, CD measurements were also carried out in various concentrations of TFE to assess the relative abilities of Cx40CT and Cx43CT to form (or stabilize current) α-helical structures (Fig. 3, B and C). According to the 50% TFE concentration, the maximum absorption of Cx40CT is shifted from 200 nm up to 205 nm. Moreover, the weak absorption at 222 nm increased. These results suggest that the Cx40CT domain is able to form an α-helical structure. A similar result is observed with Cx43CT. The main absorption at 201 nm is shifted to 206 nm in the presence of 50% TFE, and the absorption at 222 nm increased. These results confirm the presence of α-helical structure for the Cx43CT (36) and strongly suggest that the Cx40CT domain has the ability to form α-helical structure as well.
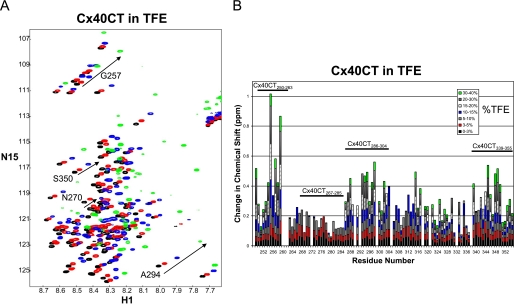
To identify if Cx40CT residues Asp-279—Ser-286 (displaying the hydrogen bond pattern from the NMR temperature coefficient experiment) were responsible for forming the α-helical structure, we collected 15N-HSQC spectra of the Cx40CT as a function of TFE concentration (Fig. 4A). The 15N-HSQC is a two-dimensional NMR experiment where each amino acid (except proline) gives one signal (or chemical shift) that corresponds to its N-H amide group. These signals are sensitive to the chemical environment, and even small changes in structure and/or dynamics can change the chemical shift of an amino acid. The addition of TFE from 3 to 40% caused a majority of the Cx40CT amino acids to shift position (e.g. Gly-257, Ala-294, and Ser-350), and a subset of amino acids that initially shifted then disappeared or “broaden beyond detection” with increasing concentrations of TFE (e.g. Asn-270). Four unique regions were identified when each amino acid was plotted against its change in chemical shift (Fig. 4B). Three areas had a cluster of residues that shifted the greatest in the presence of TFE (Cx40CT250–263, Cx40CT286–304, and Cx40CT339–355), and one area had a cluster of residues that shifted and then disappeared (Cx40CT267–285). Synthetic peptides for each of these Cx40CT regions were generated and analyzed by CD.
FIGURE 4.
15N-HSQC spectra of the Cx40CT in the presence of TFE. A, the control spectrum, Cx40CT-only (black), has been overlapped with spectra obtained when the Cx40CT was in the presence of 5% (red), 15% (blue), and 40% (green) TFE. Spectra for the Cx40CT in the presence of 3, 10, 20, and 30% were also collected but not shown due to the overlap of the peaks. B, each Cx40CT residue was plotted against their change in chemical shift as a function of TFE concentration. The four major affected areas have been labeled, and a representative residue for each area is displayed in panel A.
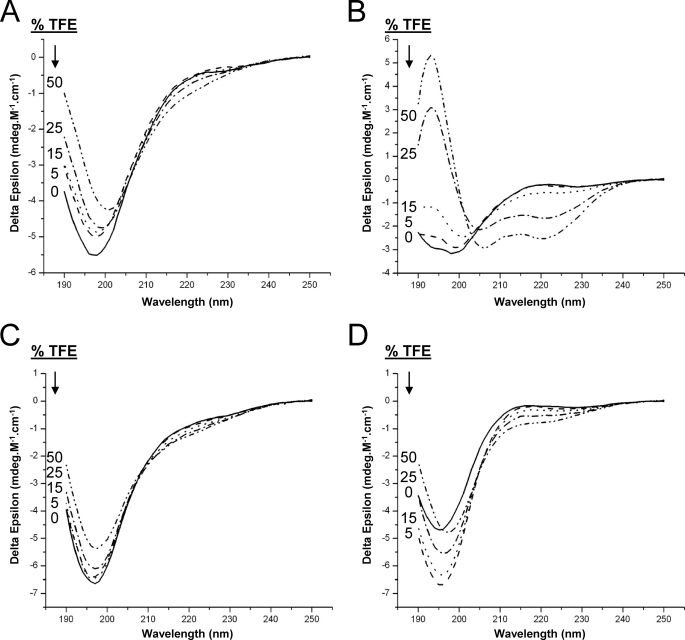
Similar to the wild-type Cx40CT, CD data were collected for each peptide in the absence and presence of various concentrations of TFE (Fig. 5). Clearly, only peptide Cx40CT267–285 was able to form an α-helical structure (Fig. 5B), suggesting that this region has a propensity to form an α-helical structure.
FIGURE 5.
CD spectra of Cx40CT peptides in the presence of TFE. CD spectra of Cx40CT peptides (A) Cx40CT250–263, Cx40CT267–285 (B), Cx40CT286–304 (C), and Cx40CT339–355 (D) were collected in the absence and presence of varying concentrations of TFE (% displayed in each panel).
Cx40CT Oligomerization
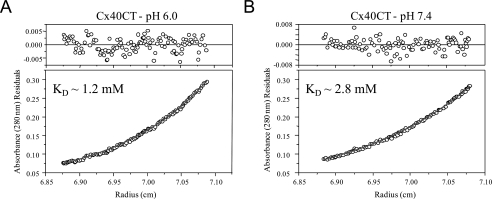
Previously we identified that acidification causes Cx43CT dimer formation in vitro (24). To assess the possibility of pH-dependent dimerization of Cx40CT, solutions of purified, recombinant Cx40CT were analyzed at pH 6.0 and 7.4 by sedimentation equilibrium. This method also allows us to accurately quantify the stoichiometry of oligomerization at both pH values. Plots of optical density at equilibrium as a function of radius at pH 6.0 and 7.4 at a rotor speed of 24,000 rpm are shown in Fig. 6, A and B, respectively. Data were also collected at 25,000 and 26,000 rpm for both pH values (data not shown) and used to analyze the stoichiometry of oligomerization. Each plot was best fit by a function derived from a self-association model to determine the fraction of protein in specific oligomeric states. A monomer-dimer model showed very good convergence, as demonstrated by the minimum deviation seen in the residuals (Fig. 6, top plot on each panel) and a weighted variance approaching unity (data not shown). The results obtained from the monomer-dimer model were used to calculate the dissociation constant (KD) of Cx40CT oligomerization at both pH values (see the inset in Fig. 6, A and B). In contrast, fits of data to single component (i.e. non-oligomeric), monomer-dimer-trimer, and monomer-dimer-tetramer models had weighted variance values significantly greater than one (data not shown). The results for the monomer-dimer model were used to calculate the fraction of total protein that existed in the dimer conformation at a concentration of 70 μm (concentration used to calculate the % dimerization for the Cx43CT domain (24)). At both a pH of 6.0 and 7.4, only a small fraction of the total protein content was in the dimer conformation (5.2% and 2.3%, respectively). The small pH dependence to the process of Cx40CT dimerization is significantly less than what was observed previously for Cx43CT dimerization (pH 5.8, 86% and pH 7.5, 12% (24)).
FIGURE 6.
Characterization of the pH-dependent Cx40CT self-association by sedimentation equilibrium. Determination of Cx40CT oligomerization by sedimentation equilibrium is shown. The absorbance at 280 nm of Cx40CT in PBS buffer at pH 6.0 (A) and pH 7.4 (B) at equilibrium (24,000 rpm) is shown as a function of radius. The solid lines are the theoretical curves. The KD for Cx40CT dimerization at both pH values is indicated in each box.
Interaction between the Cx40CT and Cx43CT Domains
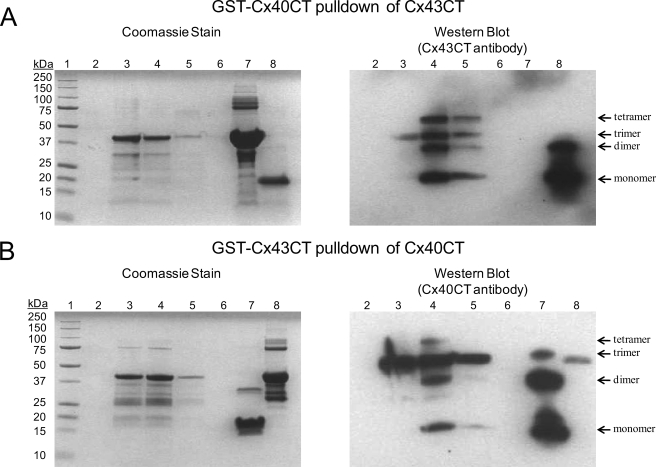
The existence of heteromeric Cx40/Cx43 connexons and the ability of Cx43CT and Cx40CT to form dimers, albeit much weaker for the Cx40CT, suggest a possible interaction between these domains. To test for hetero-CT dimerization, we used an in vitro GST-tagged pulldown system, NMR, and SPR experiments. Purified GST-tagged Cx43CT or Cx40CT bound to glutathione beads was incubated with pure Cx40CT and Cx43CT, respectively, for 1 h and washed, and the proteins were eluted with 100 mm glutathione. Equal volumes of sample were loaded onto Tris-Tricine SDS-acrylamide gels and either stained with Coomassie Blue or further analyzed by Western blot (Fig. 7). For the Western blot of the GST-Cx40CT pulldown of Cx43CT using a Cx43CT antibody (Fig. 7A), as expected lanes containing beads alone (lane 2), beads and GST-Cx40CT (lane 3), the blank lane (lane 6), and pure GST-Cx40CT (lane 7) were blank. However, the first and second elution washes with glutathione (lanes 4 and 5) contained Cx43CT, indicating a direct interaction with the Cx40CT. Interestingly, purified Cx43CT (positive control lane 8) contained a major monomeric band as well as a dimeric band, whereas the elution wash (lanes 4 and 5) also contained trimer and tetramer bands. In the reverse orientation, unfortunately the Cx40CT anti-body had an affinity for the Cx43CT as demonstrated in lanes 3 (beads and GST-Cx43CT) and 8 (purified GST-Cx43CT) (Fig. 7B). Although nonspecific binding occurred, the presence of the Cx40CT band in lanes 4 (elution wash 1) and 5 (elution wash 2) indicates a direct interaction with the Cx43CT. Therefore, in both orientations the in vitro binding assay identified a direct interaction between the Cx43CT and Cx40CT.
FIGURE 7.
Demonstrating a direct interaction between the Cx43CT and Cx40CT domains using a GST pulldown assay. A and B, the ProFound pulldown GST Protein:Protein Interaction kit (Thermo Scientific) was used to test for the Cx43CT/Cx40CT interaction. The proteins were resolved on 10% Tris-Tricine SDS-acrylamide gels and analyzed using Coomassie Blue Stain and an anti-Cx43CT or anti-Cx40CT antibody (labeled in each panel). A, shown is GST-Cx40CT pulldown of Cx43CT. Lane 1, molecular weight standard; lane 2, glutathione resin; lane 3, glutathione resin plus GST-Cx40CT; lane 4, GST-Cx40CT plus Cx43CT (elution 1); lane 5, GST-Cx40CT plus Cx43CT (elution 2); lane 6, blank; lane 7, pure GST-Cx40CT (negative control); lane 8, pure Cx43CT (positive control). B, shown is GST-Cx43CT pulldown of Cx40CT. Lane 1, molecular weight standard; lane 2, glutathione resin; lane 3, glutathione resin plus GST-Cx43CT; lane 4, GST-Cx43CT plus Cx40CT (elution 1); lane 5, GST-Cx43CT plus Cx40CT (elution 2); lane 6, blank; lane 7, pure Cx40CT (positive control); lane 8, pure GST-Cx43CT (negative control).
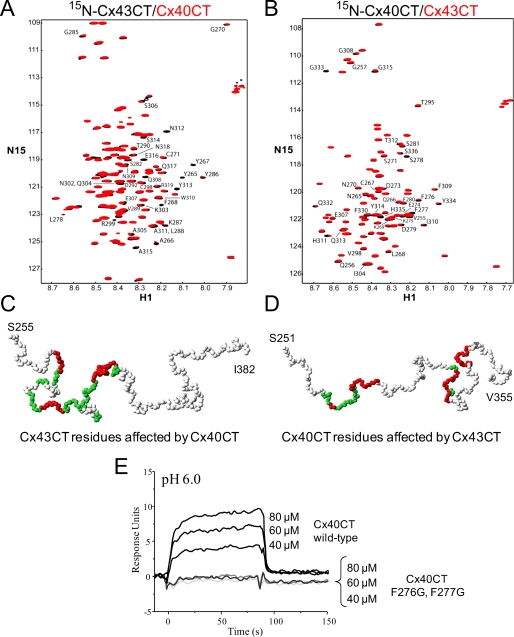
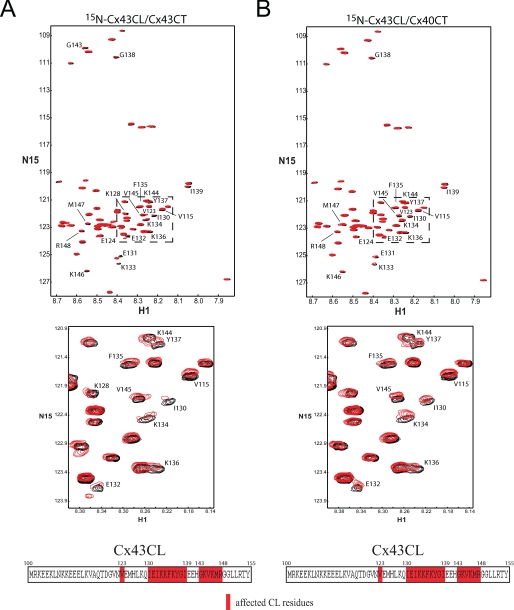
NMR experiments were performed with unlabeled Cx40CT or Cx43CT titrated into a PBS buffer solution (pH 6.0) containing 15N-Cx43CT or 15N-Cx40CT, respectively, and 15N-HSQC spectra were acquired (Fig. 8, A and B). We attempted to calculate the dissociation constant (KD) for the Cx43CT/Cx40CT interaction by holding the concentration of the 15N-labeled CT domain constant (100 μm) and titrating the unlabeled CT domain from 100 μm to 3.2 mm; however, we could not reach saturation due to the solubility of the CT domains. We extrapolated the curves from both CT points of view (decreasing signal intensity versus CT concentration), and the titration data afforded a KD of ∼4 mm. The spectra displayed in Fig. 8, A and B, are those of the 1:32 molar ratio because this ratio generated the greatest change in chemical shifts. The percent occupancy of the 15N-labeled CT at the 1:32 ratio using a KD of ∼4 mm is only ∼45% (as apposed to <1.0% at a 1:1 molar ratio). The addition of either the unlabeled Cx40CT or Cx43CT to their 15N-labeled counterpart affected a number of resonance peaks throughout the length of both domains.
FIGURE 8.
Characterizing the interaction between the Cx43CT and Cx40CT domains. A and B, Cx40CT/Cx43CT interaction by NMR is shown. 15N-HSQC spectra were collected for the 15N-Cx43CT in the presence of unlabeled Cx40CT (A) and visa versa (B) at a 1:32 molar ratio. This experiment allowed for detection of the Cx43CT and Cx40CT residues that were structurally modified when in the presence of each other. The affected Cx43CT and Cx40CT residues have been labeled. C and D, shown is a three-dimensional model of the Cx43CT and Cx40CT residues labeled according to their degree of disappearance in the presence of each other (red greater than green). E, Cx40CT/Cx43CT interaction by SPR is shown. Sensograms obtained after various concentrations of Cx40CT wild-type or the Cx40CTF276G,F277G mutant at pH 6.0 were added to a chip where the Cx43CT domain had been immobilized. The concentrations of Cx40CT wild-type and mutant added to the chip have been labeled.
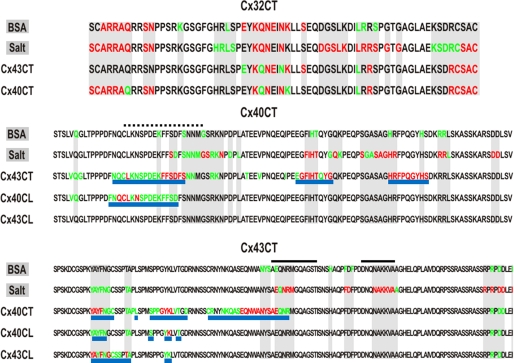
The low affinity for the Cx40CT/Cx43CT interaction and the need for large concentrations of ligand raises the question of whether there are nonspecific effects/artificial associations between the CT domains. To address this issue, we used purified Cx32CT, an isoform known not to make heteromers with Cx40 or Cx43, to test if it can interact with both the Cx43CT and Cx40CT domains. Additionally, we used bovine serum albumin (BSA) as a control to test for nonspecific binding to the CT domains and 2× PBS to test if changes in salt concentration affect the CT domains (Fig. 9). Surprisingly, BSA, salt, Cx43CT, and Cx40CT all affected the Cx32CT. What is important is that when the Cx32CT residues affected by BSA and salt are highlighted (Fig. 9, top panel, gray), they encompass all of the Cx32CT residues that are affected by the Cx43CT or Cx40CT. Thus, the data suggest that Cx43CT and Cx40CT non-specifically affect the Cx32CT at these high concentrations (i.e. 1:32 molar ratio). When the same binding experiments were performed from the Cx43CT and Cx40CT point of view and when the residues affected by BSA and salt were highlighted (Fig. 9, middle and lower panel; gray), additional residues were affected that are specific for the Cx43CT/Cx40CT interaction. We consider these residues (Fig. 8, C and D, and Fig. 9, underlined in blue) the ones affected by the interaction. The Cx43CT domains affected by the Cx40CT were Tyr-265—Cys-271, Ser-282—Asp-292, and Cys-298—Arg-319, and the Cx40CT domains affected by the Cx43CT were Asn-265—Ser-281, Glu-307—Gly-315, and His-328—Ser-336. Each CT domain had three strongly affected regions.
FIGURE 9.
Summary of the Cx32CT, Cx40CT, and Cx43CT residues affected by BSA and salt concentration. Displayed is the amino acid sequence for the Cx32CT (top), Cx40CT (middle), and the Cx43CT (bottom) domains. On the left side of each sequence is the ligand titrated into the CT domain, and the affected residues have been highlighted (red greater than green). Gray-shaded regions indicate the CT residues that are affected by BSA and salt. After analyzing the Cx40CT and Cx43CT residues affected by BSA and salt, underlined in blue are the CT residues affected by the presence of the indicated CT or CL ligand. Black lines are the Cx43CT residues that form an α-helical structure. Black dashed lines are the Cx40CT residues determined from the CD/TFE experiment to form α-helical structure.
The observation of an association between Cx43CT/Cx40CT was also confirmed by SPR (Fig. 8E). Here, we also used a Cx40CT construct where both Phe-276 and Phe-277 were mutated to a glycine residue. These residues are strictly conserved among all species, suggesting their importance, and these two residues were strongly affected by the interaction with the Cx43CT domain. The Cx43CT domain was immobilized on a CM-5 chip, and sensograms were obtained after injection of various concentrations of Cx40CT wild type or the Cx40CTF276G,F277G mutant at pH 7.4 and 6.0. Clearly, the Cx40CT wild type interacted with the Cx43CT domain, whereas mutation of Cx40CT residues Phe-276 and Phe-277 completely abolished this interaction. These data support the association of these two domains, as was observed in the GST pulldown assay and NMR.
Hetero Interactions between Cytoplasmic Domains of Cx40 and Cx43
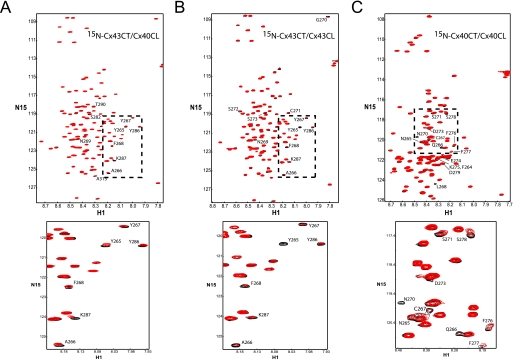
Previous electrophysiological studies in Xenopus oocytes have suggested that the CT domain of Cx43 or Cx40 could regulate the channel formed by a Cx43 construct missing its CT domain (7). We identified previously that the Cx43CT can interact with the Cx43CL domain in a pH-dependent manor (23, 25), supporting the particle-receptor hypothesis for Cx43 channel gating. Here, we used NMR to test if the particle-receptor hypothesis applies to Cx40 as well as when the two domains (i.e. CT and CL) are from different connexin isoforms. NMR experiments were performed with unlabeled Cx40CL or Cx43CL titrated into a PBS buffer solution (pH 6.0) containing 15N-Cx43CT or 15N-Cx40CT, and 15N-HSQC spectra were acquired (Fig. 10). The addition of the Cx40CL or Cx43CL domains affected a number of similar, but not identical resonance peaks between Cx43CT residues Tyr-265 and Thr-290 (summary in Fig. 9; a three-dimensional model displaying the Cx43CT residues affected is provided in supplemental Fig. 3, A and B). Cx43CT residues Arg-376 and Asp-378 are not included because these residues were sensitive to BSA and salt. The Cx43CL caused a greater decrease in Cx43CT signal intensity than the Cx40CL (e.g. Tyr-265 and Thr-275), suggesting a greater binding affinity for the Cx43CT/Cx43CL interaction. We would like to mention that we have redefined the Cx43CT binding domain previously described using the Cx43CL and Cx43CL peptides (23). The positions of the Cx43CT resonance peaks are extremely sensitive to changes in salt concentration as described above as well as trifluoroacetic acid.4 Trifluoroacetic acid is commonly used in the manufacturing process to release synthesized peptides from solid-phase resins and/or used during reversed-phase high performance liquid chromatography purification of peptides. Our previous study used a synthetic Cx43CL peptide for the NMR binding studies with the Cx43CT.
FIGURE 10.
Characterizing the interaction between the CT and CL domains of Cx43 and Cx40 from the CT point of view. These experiments allowed for detection of the regions of Cx43CT or Cx40CT that were structurally modified when in the presence of a binding domain from the molecular partners Cx43CL and Cx40CL. The Cx43CT was titrated with the Cx40CL (panel A) or Cx43CL (panel B) domains to a 1:35 molar ratio. The Cx40CT was titrated with the Cx40CL domain (panel C) to a 1:35 molar ratio. 15N-HSQC spectra for Cx43CT or Cx40CT alone (black) have been overlaid with spectra obtained in the presence of the Cx40CL or Cx43CL domains (red). Below each panel is a close-up of resonance peaks from the box in each panel.
The addition of the Cx40CL domain affected the resonance peaks between Cx40CT residues Phe-264 and Asp-279 (see the summary in Fig. 9; a three-dimensional model displaying the Cx40CT residues affected is provided in supplemental Fig. 3C). Interestingly, both CL domains affected a similar area on the amino terminus of the CT domains. In the reverse orientation, unlabeled Cx43CT and Cx40CT not only interact with 15N-labeled Cx43CL but affect identical Cx43CL residues (Fig. 11, A and B). Again, we used BSA as a control to test for nonspecific binding to the Cx43CL domain and 2× PBS to test if changes in salt concentration affect the Cx43CL domain (supplemental Fig. 4). Both BSA and 2× PBS affected most of the COOH-terminal Cx43CL residues; however, the pattern is not the same as when in the presence of the CT domains. For example, BSA and salt strongly affected Cx43CL residues Glu-124—Gln-129 and Met-147—Tyr-155, respectively, yet in the presence of the Cx43CT and Cx40CT domains, these residues were unaffected. Thus, although the Cx43CL residues affected by the CT domains overlap with those affected by BSA and salt, the difference in pattern suggests Cx43CL residues Val-123, Ile-130—Ile-139, and Gly-143—Arg-148 are the residues affected due to an interaction with the CT domains. The Cx43CT caused a greater decrease in Cx43CL signal intensity than the Cx40CT, again indicating a greater binding affinity for the Cx43CT/Cx43CL interaction over the hetero-connexin CT/CL interaction. Overall, these studies provide evidence that interactions between domains from different connexins could be critical for the regulation of heteromeric channels.
FIGURE 11.
Characterizing the interaction between the CT and CL domains of Cx43 and Cx40 from the Cx43CL point of view. These experiments allowed for detection of the regions of Cx43CL that were structurally modified when in the presence of molecular partners Cx43CT and Cx40CT. The Cx43CL was titrated with the Cx43CT (panel A) or Cx40CT (panel B) domains to a 1:35 molar ratio. 15N-HSQC spectrum for Cx43CL alone (black) has been overlaid with spectra obtained in the presence of the Cx43CT or Cx40CT domains (red). Below each panel is a close-up of resonance peaks from the box in each panel and a summary of the Cx43CL residues affected by the Cx43CT and Cx40CT domains.
DISCUSSION
We have biophysically characterized the structure of the CT domain from Cx40 by the use of NMR, FTIR, and CD. Our data indicate that the Cx40CT domain alone exists as an intrinsically disordered protein; however, one region has a propensity to form an α-helical structure. We have also used sedimentation equilibrium to characterize the process of pH-dependent Cx40CT self-association. Our data suggest that Cx40CT has the ability to dimerize, albeit weakly, with a slightly higher affinity under acidic conditions. We further showed by a GST pulldown assay, NMR, and SPR that the Cx40CT domain can interact with the Cx43CT domain. Finally, we demonstrated by NMR that the CT domains of Cx40 and Cx43 can also interact with the CL domains of Cx40 and Cx43 (and vice versa, respectively). These studies represent the first structural characterization of a connexin domain when integrated in a complex with another connexin domain and provide insight into the molecular mechanisms involved in the chemical regulation of homo- and heteromeric gap junction channels.
Previous studies have shown, functionally and biochemically, that Cx40 and Cx43 form heteromeric connexons when expressed in the same cell, and these connexons display different biophysical properties when compared with their homomeric forms (12, 14–16, 18–20). For example, the CT domain of Cx43 enhanced the pH sensitivity of a Cx40 CT-truncated mutant and vice versa (7), indicating heterodomain interactions (i.e. CT and CL) involving different connexins (i.e. Cx43 and Cx40) within a connexon are more efficient than heterodomain interactions by the same connexin at closing the channel. The Delmar laboratory (7) has previously demonstrated that pH gating of both Cx43 and Cx40 follows the particle-receptor (or “ball-and-chain”) model, and our combined studies have identified that the CL domain is the “receptor” for the CT domain acting as the gating “particle” to close Cx43 gap junction channels (23, 25, 37). Here, our studies further support the particle-receptor model for pH gating by the observation of a direct interaction between the CL and CT domains of Cx40. In addition, we have also observed a direct interaction between the CT of Cx43 and the CL of Cx40. Therefore, one component responsible for this synergism could be the interaction between the CT domain of one connexin and the CL domain of another connexin. Our work presented here also suggests that this synergism may also be a result of the interaction between the CT domain of one connexin and the CT domain of the other. The greatest change in pH sensitivity was observed when Cx40 and Cx43 were co-expressed as compared with truncation mutations or chimeras (7, 38). In the later, only homo-CT oligomerization could occur, whereas with co-expression, hetero-CT oligomerization can occur. It is tempting to speculate that hetero-oligomerization between Cx43CT and Cx40CT is responsible for the greater sensitivity.
Comparison of the Cx40CT and Cx43CT Structures
The Cx43CT structure was determined to be mostly random-coiled, with two short helical domains (24). In contrast, the Cx40CT structure is entirely random-coiled; however, one region (Cys-267—Gly-285) has a propensity to form an α-helical structure. Although the Cx43CT α-helical domains were able to be modeled from the NOE restraints, not all of the expected connectivities (e.g. αN(i,i+4)) found in an α-helical structure were identified in the NOESY spectra. This suggests that the Cx43CT α-helical domains are “dynamic.” From the structures of these CT domains arises the question as to what is the functional significance of the random-coiled and α-helical domains?
Functional Significance of the Random-coiled Domains
In general, random-coiled or intrinsically disordered domains have been identified as playing important roles in molecular recognition, molecular assembly, and protein modifications (39). In the case of membrane proteins, like Cx40 and Cx43, intrinsically disordered domains seem to play an important role in cell signaling events, acting as components of macromolecular processes and substrates for regulatory events such as phosphorylation (40). We speculate that a disordered CT allows the connexin molecule to be a point of convergence for alternative regulatory pathways. Multiple alternative interactions of low affinity (even if of high specificity) allow a single molecule to rapidly transition from one molecular partner to another, thus acting as a “switchboard” for adapting the properties of cell communication to the environment prevalent in the intracellular space (e.g. c-Src and ZO-1 (36, 41–43)).
Additionally, six disordered CT domains in a connexon may regulate channel activity by acting as one entropic bristle domain. The entropic bristle domain is not a structurally stable domain with a folded state but the result of thermal motion that acts to exclude larger molecules while allowing water and small solutes to move freely through the channel (44). This might be expected to occur after scaffolding proteins such as ZO-1 and α/β-tubulin are released from the CT domain. The CT domains of Cx40 and Cx43 are not alone in their structural characteristics. Analysis of genomic sequence data strongly suggested that there is a high prevalence of intrinsically disordered domains. Indeed, an estimated 41% of human membrane proteins, including connexins, glutamate (N-methyl-d-aspartate) receptor subunit ϵ4, and voltage-dependent T-type calcium channel α-1G subunit, have intrinsically disordered regions with more than 30 consecutive residues, and these residues are preferentially localized at the cytoplasmic side (45).
Functional Significance of the α-Helical Domains
Previously, we showed that the Cx43CT domain is predominately in a dimer conformation under acidic conditions and that residues within the two α-helical domains are involved in the dimerization interface (23, 24). In contrast, one Cx40CT domain has a propensity to form an α-helix, and sedimentation equilibrium studies indicated only a small amount of dimerization upon acidification of the solvent. This suggests that α-helices may be important for stabilizing the CT dimer conformation under acidic conditions (i.e. more α-helix leads to a stronger binding affinity for the dimer conformation). We hypothesize that CT dimerization is part of the structural foundation for the ball-and-chain model of chemical gating of Cx43 and Cx40 gap junction channels (7, 21, 22), which stabilizes the direct intramolecular interaction between the CT and CL domains (23, 25) during acidification-induced closure of the channel. Predictions from their sequence indicate that analogous helical structures also occur in other connexin CTs (e.g. Cx46 and Cx50).
Many Cx43CT and Cx40CT residues were affected upon their interaction, including the first Cx43CT and the only “potential” Cx40CT α-helical domain as well as two previously described Cx43CT homodimerization domains (23). This suggests that hetero-oligomerization is similar but not identical to homo-oligomerization. The major difference for hetero-oligomerization is the number of residues in the amino terminus of Cx43CT that were affected versus more throughout the sequence for Cx43CT homo-oligomerization. Although the binding affinity for Cx43CT and Cx40CT homo-oligomerization is greater (17 μm (24) and 1.2 mm, respectively) than for hetero-oligomerization (∼4 mm) in vitro, the possibility of hetero-oligomerization does exist in the context of a functional gap junction channel. Using the radius of a connexon of 35 Å and a Cx43CT (or Cx40CT) height of 100 Å (46), this places the effective local concentration of 6 Cx43CT particles at the mouth of an assembled connexon to be ∼25 mm. Taken alone, this would push the percent Cx43CT and Cx40CT homodimerization to 99.9 and 95.4%, respectively, and heterodimerization to 86.2%. Interestingly, antibody-recognized bands of the approximate dimeric mobility have also been seen for Cx40 (47) and for Cx43 (48–50), thus supporting the possibility that Cx40 homo- and Cx43/Cx40 hetero-CT dimerization may occur in the setting of an assembled channel.
We have recently characterized the Cx43CT when attached to the fourth transmembrane domain (residues Gly-178—Ile-382) and determined the α-helical content for this construct to be 46% as compared with 5% for the soluble Cx43CT (51). With the predicted length of the fourth transmembrane domain to be ∼10% that of the total number of amino acids, this would suggest that the α-helices extend out from the TM4 into the solution and/or the length of the two existing α-helices has increased. If the TM4-Cx40CT follows the same observation, we may expect residues Cys-267—Gly-285 to form a stable α-helix. Therefore, we speculate that the increased α-helical content would increase the binding affinity for homo- and hetero-CT dimerization and the CT/CL interaction in vivo. Additionally, the fact that these CTs are not free in solution but oriented with fixed positions due to their attachment to the membrane would suggest that homo- and heterotrimer conformations (i.e. a “dimer of trimer”) cannot be ruled out as a possible physiologically significant quaternary structure.
Functional Significance of the CT/CL Interaction
We have previously reported that Cx43CT interacts with a peptide corresponding to the second half of the Cx43CL (amino acids 119–144) and that although a peptide of region 100–118 does not directly bind to Cx43CT, the presence of this sequence is important for an increased binding affinity with the Cx43CT domain (23, 25). Using a longer Cx43CL construct (amino acids 100–155) to better maintain the functional integrity of the Cx43CL domain, we have confirmed that Cx43CT interacts with many residues within the second half of the Cx43CL. Our NMR studies presented here have also redefined the location of the Cx43CL interaction on the Cx43CT. The addition of the Cx43CL polypeptide had significant effects on the position of resonance peaks corresponding to Cx43CT amino acids Tyr-265—Ala-276 and Tyr-286—Lys-287. The previously described Cx43CT regions affected by the Cx43CL (residues Asn-343—Lys-346 and Arg-376—Asp-379 (23)) were the result of high concentrations of salt and trifluoroacetic acid from the peptide synthesis. Not only did Cx40CL affect a similar location of Cx43CT as the Cx43CL, but the Cx40CL affected a similar location on the Cx40CT (i.e. amino terminus). In contrast to our previous report (23), the location of the Cx43CT residues involved in the Cx43CL are closer to the membrane and are, thus, more juxtaposed in space with the Cx43CL. The location of the CT/CL interaction is consistent with studies from the Delmar laboratory (52) showing that Cx43CT residues Gly-261—Asn-300 are essential components of the pH-dependent “gating particle,” which is responsible for acidification-induced uncoupling of Cx43-expressing cells.
In summary, we have biophysically characterized the Cx40CT domain alone and in complex with three molecular partners, the CT and CL domains from Cx43 and the CL domain of Cx40. The Cx40CT is intrinsically disordered, with one region having a propensity to form an α-helix and the ability to dimerize. We have also shown that the Cx40CT domain interacts with the Cx40CL domain, consistent with the particle-receptor model for pH-gating of Cx40 channels. Moreover, we have observed the direct interaction and characterized the binding interface between the CT domains of Cx40 and Cx43 as well as the CL domain from one connexin with the CT domain from the other connexin. Our results suggest that despite their differences in primary sequence, the CT and CL domains of Cx40 and of Cx43 can reach a similar conformation, thus allowing for interaction with a common binding domain. Whether these hetero-CT and hetero-CT/CL interactions participate in the regulation of heteromeric channels remains to be determined.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM072631 and HL039707 (USPHS). This work was also supported by American Heart Association Grant 0560050Z, the Nebraska Research Initiative funding for the Nebraska Center for Structural Biology, and Eppley Cancer Center Support Grant P30CA036727.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
The atomic coordinates and structure factors (code 2k7m) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. Spagnol, F. Kieken, S. M. Taffet, M. Delmar, and P. L. Sorgen, unpublished data.
- CL
- cytoplasmic loop
- CT
- carboxyl terminus
- Cx43
- connexin43
- Cx40
- connexin40
- FTIR
- Fourier transform infrared
- HSQC
- heteronuclear single-quantum correlation
- PBS
- phosphate-buffered saline
- SPR
- surface plasmon resonance
- TFE
- trifluoroethanol
- GST
- glutathione S-transferase
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BSA
- bovine serum albumin.
REFERENCES
- 1.Bernstein S. A., Morley G. E. (2006) Adv. Cardiol. 42, 71–85 [DOI] [PubMed] [Google Scholar]
- 2.Delmar M. (2000) J. Cardiovasc. Electrophysiol. 11, 118–120 [DOI] [PubMed] [Google Scholar]
- 3.Dhein S. (2006) Adv. Cardiol. 42, 198–212 [DOI] [PubMed] [Google Scholar]
- 4.Hervé J. C., Dhein S. (2006) Adv. Cardiol. 42, 107–131 [DOI] [PubMed] [Google Scholar]
- 5.Spray D. C., Bennett M. V. (1985) Annu. Rev. Physiol. 47, 281–303 [DOI] [PubMed] [Google Scholar]
- 6.Francis D., Stergiopoulos K., Ek-Vitorín J. F., Cao F. L., Taffet S. M., Delmar M. (1999) Dev. Genet. 24, 123–136 [DOI] [PubMed] [Google Scholar]
- 7.Stergiopoulos K., Alvarado J. L., Mastroianni M., Ek-Vitorin J. F., Taffet S. M., Delmar M. (1999) Circ. Res. 84, 1144–1155 [DOI] [PubMed] [Google Scholar]
- 8.Bukauskas F. F., Bukauskiene A., Bennett M. V., Verselis V. K. (2001) Biophys. J. 81, 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saffitz J. E., Yamada K. A. (2000) Cardiovasc. Res. 45, 807–809 [DOI] [PubMed] [Google Scholar]
- 10.Peters N. S., Coromilas J., Severs N. J., Wit A. L. (1997) Circulation 95, 988–996 [DOI] [PubMed] [Google Scholar]
- 11.Koval M. (2006) Trends Cell Biol. 16, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He D. S., Jiang J. X., Taffet S. M., Burt J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6495–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Veen T. A., van Rijen H. V., Jongsma H. J. (2006) Adv. Cardiol. 42, 18–40 [DOI] [PubMed] [Google Scholar]
- 14.Valiunas V., Gemel J., Brink P. R., Beyer E. C. (2001) Am. J. Physiol. Heart Circ. Physiol. 281, H1675–H1689 [DOI] [PubMed] [Google Scholar]
- 15.Gu H., Ek-Vitorin J. F., Taffet S. M., Delmar M. (2000) Circ. Res. 86, 1100. [PubMed] [Google Scholar]
- 16.Elenes S., Rubart M., Moreno A. P. (1999) J. Cardiovasc. Electrophysiol. 10, 990–1004 [DOI] [PubMed] [Google Scholar]
- 17.Valiunas V., Weingart R., Brink P. R. (2000) Circ. Res. 86, E42–E49 [DOI] [PubMed] [Google Scholar]
- 18.Burt J. M., Fletcher A. M., Steele T. D., Wu Y., Cottrell G. T., Kurjiaka D. T. (2001) Am. J. Physiol. Cell Physiol. 280, C500–C508 [DOI] [PubMed] [Google Scholar]
- 19.Cottrell G. T., Burt J. M. (2001) Am. J. Physiol. Cell Physiol. 281, C1559–C1567 [DOI] [PubMed] [Google Scholar]
- 20.Cottrell G. T., Wu Y., Burt J. M. (2002) Am. J. Physiol. Cell Physiol. 282, C1469–C1482 [DOI] [PubMed] [Google Scholar]
- 21.Morley G. E., Taffet S. M., Delmar M. (1996) Biophys. J. 70, 1294–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anumonwo J. M., Taffet S. M., Gu H., Chanson M., Moreno A. P., Delmar M. (2001) Circ. Res. 88, 666–673 [DOI] [PubMed] [Google Scholar]
- 23.Hirst-Jensen B. J., Sahoo P., Kieken F., Delmar M., Sorgen P. L. (2007) J. Biol. Chem. 282, 5801–5813 [DOI] [PubMed] [Google Scholar]
- 24.Sorgen P. L., Duffy H. S., Spray D. C., Delmar M. (2004) Biophys. J. 87, 574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duffy H. S., Sorgen P. L., Girvin M. E., O'Donnell P., Coombs W., Taffet S. M., Delmar M., Spray D. C. (2002) J. Biol. Chem. 277, 36706–36714 [DOI] [PubMed] [Google Scholar]
- 26.Bouvier D., Kieken F., Sorgen P. L. (2007) Biomol. NMR Assign. 1, 155–157 [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 29.Kay L. E., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 30.Baxter N. J., Williamson M. P. (1997) J. Biomol. NMR 9, 359–369 [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 32.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 33.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 34.Lees J. G., Smith B. R., Wien F., Miles A. J., Wallace B. A. (2004) Anal. Biochem. 332, 285–289 [DOI] [PubMed] [Google Scholar]
- 35.Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorgen P. L., Duffy H. S., Sahoo P., Coombs W., Delmar M., Spray D. C. (2004) J. Biol. Chem. 279, 54695–54701 [DOI] [PubMed] [Google Scholar]
- 37.Seki A., Duffy H. S., Coombs W., Spray D. C., Taffet S. M., Delmar M. (2004) Circ. Res. 95, e22–e28 [DOI] [PubMed] [Google Scholar]
- 38.Gu H., Ek-Vitorin J. F., Taffet S. M., Delmar M. (2000) Circ. Res. 86, E98–E103 [PubMed] [Google Scholar]
- 39.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradoviæ Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 40.Dunker A. K., Obradovic Z. (2001) Nat. Biotechnol. 19, 805–806 [DOI] [PubMed] [Google Scholar]
- 41.Toyofuku T., Akamatsu Y., Zhang H., Kuzuya T., Tada M., Hori M. (2001) J. Biol. Chem. 276, 1780–1788 [DOI] [PubMed] [Google Scholar]
- 42.Duffy H. S., Ashton A. W., O'Donnell P., Coombs W., Taffet S. M., Delmar M., Spray D. C. (2004) Circ. Res. 94, 215–222 [DOI] [PubMed] [Google Scholar]
- 43.Bouvier D., Kieken F., Kellezi A., Sorgen P. L. (2008) Cell Commun. Adhes. 15, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoh J. H. (1998) Proteins 32, 223–228 [PubMed] [Google Scholar]
- 45.Minezaki Y., Homma K., Nishikawa K. (2007) J. Mol. Biol. 368, 902–913 [DOI] [PubMed] [Google Scholar]
- 46.Unger V. M., Kumar N. M., Gilula N. B., Yeager M. (1999) Science 283, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 47.Matesic D., Tillen T., Sitaramayya A. (2003) Cell Biol. Int. 27, 89–99 [DOI] [PubMed] [Google Scholar]
- 48.VanSlyke J. K., Musil L. S. (2000) Methods 20, 156–164 [DOI] [PubMed] [Google Scholar]
- 49.Hossain M. Z., Murphy L. J., Hertzberg E. L., Nagy J. I. (1994) J. Neurochem. 62, 2394–2403 [DOI] [PubMed] [Google Scholar]
- 50.Roger C., Mograbi B., Chevallier D., Michiels J. F., Tanaka H., Segretain D., Pointis G., Fenichel P. (2004) J. Pathol. 202, 241–246 [DOI] [PubMed] [Google Scholar]
- 51.Kellezi A., Grosely R., Kieken F., Borgstahl G. E., Sorgen P. L. (2008) Protein Expr. Purif. 59, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ek-Vitorín J. F., Calero G., Morley G. E., Coombs W., Taffet S. M., Delmar M. (1996) Biophys. J. 71, 1273–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.