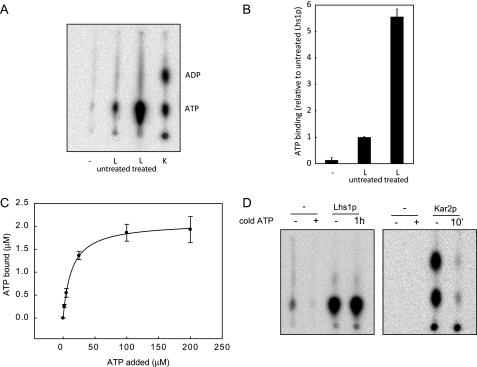
FIGURE 2.
GdnHCl-EDTA-treated Lhs1p exhibits efficient nucleotide binding activity. A, untreated Lhs1p (L), GdnHCl-treated Lhs1p or Kar2p (K) were incubated with [α-32P]ATP for 30 min at room temperature. After removal of free nucleotides by gel filtration binding, bound nucleotides were analyzed by thin layer chromatography. Consistent with previous observations (10), Lhs1p does not hydrolyze [α-32P]ATP like the canonical Hsp70 Kar2p. B, [α-32P]ATP·Lhs1p complexes were captured onto polyvinylidene difluoride filters (Millipore), followed by removal of unbound nucleotides by three washes with ice-cold ATPase assay buffer. The amount of bound [α-32P]ATP was analyzed by scintillation counting. The mean ± S.D. (error bars) from three experiments is shown. C, 3.5 μm Lhs1p was incubated for 1 h at room temperature with 0–200 μm ATP, supplemented with [α-32P]ATP. Lhs1p with bound nucleotide was captured onto polyvinylidene difluoride filters as described in B, the amount of bound [α-32P]ATP was analyzed by scintillation counting, and binding curves were generated using a one-site saturation binding model. Error bars represent the mean of two experiments that were each performed in triplicate. D, GdnHCl-treated Lhs1p or Kar2p were preincubated with [α-32P]ATP for 3 h or 30 min, respectively, a more than 1000-fold excess of cold ATP was added and the proteins were incubated for an additional hour (Lhs1p) or 10 min (Kar2p). After removal of free nucleotides by gel filtration, bound nucleotides were analyzed by thin layer chromatography.