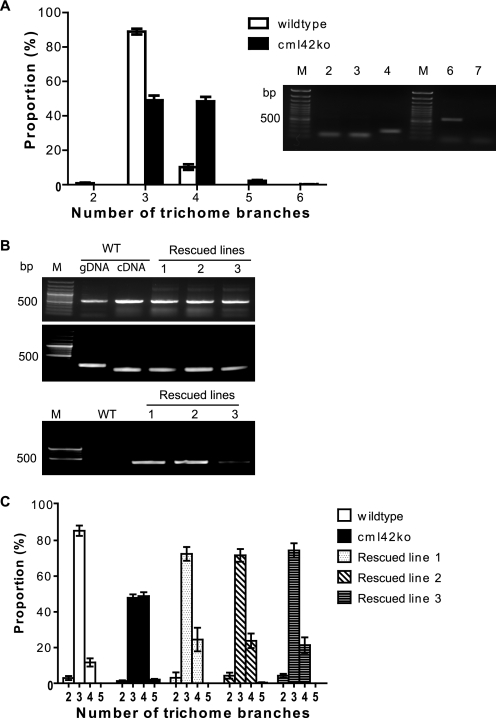
FIGURE 10.
cml42 knockout plants have increased trichome branching and can be rescued by the wild-type CML42 gene. A, number of trichome branches (mean ± S.E.) of wild-type (open bars) compared with cml42 knockout (black bars) plants. Asterisks indicate statistically significant differences (t test, p < 0.005) between wild-type and cml42 knockout plants. Inset shows RT-PCR detection of CML42 transcript in leaf tissue. Actin controls are shown in lanes 2 and 3 for wild-type and cml42 knockout plants, respectively; lane 4 shows actin amplified from genomic DNA (intron present) derived from a cml42 knockout plant; lanes 6 and 7 show CML42 transcript amplification from wild-type and cml42 knockout plants, respectively; lanes M contain DNA ladders. B, RT-PCR (top panel) was conducted to examine the presence of CML42 transcript in wild-type (WT) and three independent transgenic lines of CML42-transformed cml42 knockout plants (rescued lines 1–3). Actin was used as a control (middle panel). C, PCR was also performed (lower panel) on genomic DNA to confirm that the CML42 rescued lines were from the cml42 knockout genetic background. The presence of a T-DNA insert (primers SALK LBc1 and CML42rtR) in the endogenous CML42 gene was detected only in the rescued lines. DNA marker (M) lanes are shown on the left of each panel. C, number of trichome branches (mean ± S.E.) in wild-type plants (open bars) compared with cml42 knockout plants (solid bars) and the three independent rescued lines described in B.