Abstract
Sam68, Src associated in mitosis of 68 kDa, is a known RNA-binding protein and a signaling adaptor protein for tyrosine kinases. However, the proteins associated with Sam68 and the existence of a Sam68 complex, its mass, and regulation are, however, unknown. Herein we identify a large Sam68 complex with a mass >1 MDa in HeLa cells that is composed of ∼40 proteins using an immunoprecipitation followed by a mass spectrometry approach. Many of the proteins identified are RNA-binding proteins and are known components of a previously identified structure termed the spreading initiation center. The large Sam68 complex is a ribonucleoprotein complex, as treatment with RNases caused a shift in the molecular mass of the complex to 200–450 kDa. Moreover, treatment of HeLa cells with phorbol 12-myristate 13-acetate or epidermal growth factor induced the disassociation of Sam68 from the large complex and the appearance of Sam68 within the smaller complex. Actually, in certain cell lines such as breast cancer cell lines MCF-7 and BT-20, Sam68 exists in equilibrium between a large and a small complex. The appearance of the small Sam68 complex in cells correlates with the ability of Sam68 to promote the alternative splicing of CD44 and cell migration. Our findings show that Sam68 exists in equilibrium in transformed cells between two complexes and that extracellular signals, such as epidermal growth factor stimulation, promote alternative splicing by modulating the composition of the Sam68 complex.
Introduction
Sam68,3 the Src-associated substrate during mitosis of 68 kDa, is a known substrate for many kinases including Src family kinases (1). Sam68 harbors numerous proline- and tyrosine-rich regions that interact with Src family kinases, phospholipase Cγ1, Grb2, Nck, and Csk, in an SH3- and SH2 domain-dependent manner (2–8). Indeed Sam68 is an adaptor protein for Csk, regulating the activity of Src family kinases during cellular migration and polarization (3). Sam68 was also shown to be an adaptor protein for the protein arginine methyltransferase 1 (PRMT1) at the Hoxa9 gene during certain mixed lineage leukemia (9).
Sam68 is a hnRNP K homology-type RNA-binding protein and was termed a STAR (signal transduction activator of RNA) protein, as its tyrosine phosphorylation regulates its RNA binding activity (10, 11). hnRNP K homology-type RNA-binding proteins are known to mediate specific protein-RNA interactions (11). Sam68 associates with high affinity to UAAA RNA sequences (12), and because these sequences are often found in 3′-untranslated regions, this suggests that Sam68 may bind most mRNAs. However, Sam68 binds select mRNAs, as we recently showed that Sam68 regulates the translation of only certain mRNAs during spermatogenesis (13). Sam68 is also required for HIV infection, and Sam68 mutant proteins are able to impair the HIV reverse nuclear export pathway (14, 15) and sequester negative factor mRNAs to stress granules, a structure known to sequester ribonucleoproteins (RNPs) (16). Sam68 is known to regulate the alternative splicing of CD44 and Bcl-x (17–19) and a subset of genes during neurogenesis (20). Post-translational modifications of Sam68 regulate its alternative splicing function, and an example is the Sam68 regulation of the CD44 variable exon 5 (V5) that is MAP kinase-dependent (17). The alternative splicing of Bcl-x mRNA is also regulated by tyrosine phosphorylation of Sam68 (19). Moreover, Sam68 may be involved in flagging variable exons for the polymerase II machinery (21), and it has also been shown to regulate transcription (9, 22, 23). Sam68 knock-out mice have revealed a role for Sam68 in bone physiology as the mice display continuous bone remodeling with age and, therefore, are protected against age-induced osteoporosis (24).
In transformed cells Sam68 resides in dynamic nuclear foci termed Sam68 nuclear bodies (SNBs) (25). These foci, also called stress nuclear bodies (26), are primarily found in cancerous cell lines and are absent in normal fibroblasts (25). SNBs also contain Sam68-like mammalian family members (SLM-1 and SLM-2) which are closely related to Sam68 (25, 27) and YT521-B, a splicing factor known to associate with Sam68 (28). Moreover, we showed that specific mutations within the Sam68 hnRNP K homology-type RNA binding domain alters the localization within SNBs (25), suggesting that RNA binding is required to localize Sam68 to these structures.
Phorbol ester stimulation activates the Ras/MAP kinase pathway in a protein kinase C-dependent manner, mediating a sustained increase in cell adhesion (29). Activation of the Ras/MAP kinase pathway is also achieved with growth factors such as EGF (30, 31). EGF receptor stimulation activates a wide array of signaling pathways controlling cellular proliferation, migration, differentiation, and survival (32). EGF receptor overexpression is observed in many types of epithelial cancers associated with poor prognosis as well as in cancers with high recurrence such as lung, head, and neck, colorectal, and breast cancer (33–35). The link between EGF receptor and cancer has been pivotal for generating new drug therapies.
In the present study we identified the presence of Sam68 within two cellular complexes; that is, a large >1-Mb complex and another smaller complex with a molecular mass ranging from 200 to 450 kDa. HeLa cells harbor only the large complex, and we identified more than 40 proteins that interact with Sam68 within this complex using an immunoprecipitation/mass spectrometry approach. Treatment of HeLa cells with phorbol 12-myristate 13-acetate (PMA) or EGF induced the disassociation of Sam68 within the large complex and the appearance of Sam68 within the smaller complex. The appearance of the small Sam68 complex correlated with the ability of Sam68 to alternatively splice CD44 and regulate cell migration.
EXPERIMENTAL PROCEDURES
Gel Filtration (Fast Protein Liquid Chromatograph)
The various cell lines purchased from the American Type Culture Collection (Manassas, VA) were lysed in 1% Triton X-100, 150 mm NaCl, 20 mm Tris, pH 7.4, and 0.1 mg/ml phenylmethylsulfonyl fluoride. Cell lysates were loaded onto a Superose-6 prep grade packed in a SR 25 column. Fast protein liquid chromatograph was performed on a Liquid Chromatography Controller LCC-500 Plus from GE Healthcare. The flow rate was set at 0.2 ml/min, and 1-ml fractions were collected using a FRAC-100 fraction collector. Each fraction was precipitated in 20% trichloroacetic acid and analyzed on SDS-polyacrylamide gels. Gels were then transferred to nitrocellulose membranes and analyzed by immunoblotting. The Superose-6 column was calibrated using bovine serum albumin (66 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), thyroglobulin (669 kDa), and blue dextran (2000+ kDa), all from Sigma-Aldrich.
Mass Spectrometry
Endogenous Sam68 was immunopurified from 5 × 108 HeLa cells by using 10 μg of polyclonal anti-Sam68 antibody (AD1; Millipore) coupled to 1 mg of protein A-Sepharose (Sigma). After extensive washings with lysis buffer and 1× phosphate-buffered saline, the bound proteins were eluted with 4 m urea and then precipitated with 20% trichloroacetic acid. Precipitated proteins were resuspended in Laemmli buffer, resolved by SDS-PAGE, and revealed by Coomassie Blue R-250 staining. Strips of gel containing ∼20 proteins were excised and sent to the University of Calgary Mass Spectrometry Proteomics Facility for LC-MS/MS analysis. The LC-MS/MS analysis for protein identification was performed on either a QSTAR Pulsar i Hybrid quadrupole time-of-flight mass spectrometer (Applied Biosystems/PE Sciex) or an Agilent MSD Ion Trap XCT interfaced with an Agilent 1100 series Nano LC system. For matrix-assisted laser desorption ionization-time-of-flight analysis, bands were excised as with LC-MS/MS and sent to the University of Calgary Mass Spectrometry Proteomics Facility. Peptides resulting from proteolytic digestion were analyzed using MALDI-MS Voyager-DE STR mass spectrometer from Applied Biosystems to give a mass fingerprint. The spectrums were analyzed using the M/Z program (Knexus edition, copyright 1998–2001, Proteometrics, LLC), and the peptide mass fingerprinting was analyzed using ProteinProspector Version 4.0.7 with the SwissProt.2006.17.10 data base for protein identification.
Immunoprecipitations and Immunofluorescence
Cells were lysed in 1% Triton X-100, 150 mm NaCl, 20 mm Tris. pH 7.4. and 0.1 mg/ml phenylmethylsulfonyl fluoride. Cell lysates were incubated on ice with primary antibody for 1 h. Then 30 μl of 50% protein A-Sepharose slurry was added and incubated at 4 °C for 30 min with constant end-over-end mixing. The beads were washed 3 times with lysis buffer and once with 1× phosphate-buffered saline. Protein samples were analyzed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Immunoblotting was performed using anti-Sam68 antibodies (Millipore), anti-G3BP1 antibodies (a gift from Dr. I. Gallouzi, McGill University, Montreal, Canada), and anti-YT521-B antibodies (BD Biosciences). Goat anti-mouse or goat anti-rabbit antibodies conjugated to horseradish peroxidase secondary antibodies were purchased from ICN Pharmaceuticals (Costa Mesa, CA) and detected using the chemiluminescence (ECL) detection kit (DuPont). For immunofluorescence, cells were fixed with 1% paraformaldehyde in 1× phosphate-buffered saline for 5 min at room temperature and permeabilized with 1% Triton X-100 in phosphate-buffered saline for 5 min at room temperature, as described previously (25). Sam68 localization was followed using an anti-Sam68 antibody, and nuclear staining was obtained using 4′-6-diamidino-2-phenylindole.
RNA Interference
hS68shRNA-1 was generated by ligation of oligos (5′-GAT CCC Cga tgg agc cag aga aca agT TCA AGA Gac ttg ttc tct ggc tcc atc TTT TTG GAA A-3′ and 5′-GCT TTT CCA AAA Aga tgg agc cag aga aca agT CTC TTG Aac ttg ttc tct ggc tcc atc GGG-3′) into BglII and HindIII of pSUPER retro (OligoEngine, Seattle, WA). The underlined sequences represent the targeted sequences. Human KHDRBS1 (Sam68) siGENOME SMARTpool M-020019-00-0010 siRNA was ordered from Dharmacon (Chicago, IL), and GFP siRNA duplex was assembled with a sense sequence (5′-aacacuugucacuacuuucucuu-3′) and antisense sequence (5′-gagaaaguagugacaaguguuuu-3′) (Dharmacon).
Cell Migration Assays
For chemotactic assays, Transwell chamber filters (8-μm pore size; Costar) coated with 15 μg of collagen/ml on both sides were used. 104 cells resuspended in Dulbecco's modified Eagle's medium with 1% bovine serum albumin were added to the upper chamber. The same medium supplemented with either 10% bovine serum, 50 ng/ml EGF, or phorbol 12-myristate 13-acetate (PMA) was used as a chemoattractant in the lower chamber. Cell migration was surveyed after 3, 6, 12, and 24 h. Migratory cells on the lower membrane surface were then counted after crystal blue staining. For wound healing assays, fibronectin-coated glass coverslips were seeded with either pSUPER or Sam68sh HeLa cells. Wounds were introduced using standard plastic pipette tips on the coverslip surface. Wound healing was surveyed 16 h afterward, and the cells were fixed with paraformaldehyde, permeabilized, and then visualized by phase-contrast microscopy.
RT-PCR
Sam68 null mouse embryo fibroblasts (MEFs) were transfected using LipofectamineTM and PLUSTM reagent and HeLa with Lipofectamine 2000TM as recommended by the manufacturer (Invitrogen). Total RNA from HeLa and Sam68−/− MEFs cells was isolated in 1 ml of Trizol (Invitrogen). Four micrograms of RNA was incubated at 37 °C for 1 h with 100 pmol of oligo(dT) primer and 100 units of Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's protocol. cDNAs were analyzed by PCR using the following primer sequences of the CD44 minigene (a gift from Dr. Stefan Stamm, University of Kentucky, Lexington, KY): insulin exon primer N5Ins (AGGGATCCGCTTCCTGCCCC) and N3Ins (CTCCCGGGCCACCTCCA) and glyceraldehyde-3-phosphate dehydrogenase forward (AGCCACATCGCTCAGACAC) and reverse (GCCCAATACGACCAAATCC). All amplifications were performed using 6 μl of cDNA per reaction. After 35 cycles of the reaction (95 °C for 30 s, 62 °C for 45 s, 72 °C for 60 s), the products were separated by electrophoresis on 1.5% agarose gels.
RESULTS
Identification of Sam68 Complexes
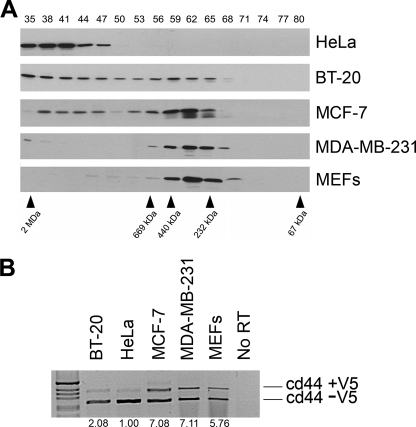
To identify the mass of a putative Sam68 complex, HeLa cellular extracts were prepared and fractionated over a Superose 6 column. The collected fractions numbered 35–80 were immunoblotted with anti-Sam68 antibodies. We observed one predominant Sam68 complex termed “the large complex” with a mass of >1 MDa (Fig. 1A, fractions 35–47). In the human breast carcinoma epithelial cell line BT-20 and the human breast adenocarcinoma epithelial cell line MCF-7, we noted the presence of both a large (>1 MDa, fractions 35–47) and a smaller Sam68 complex (Fig. 1A, ∼200–450 kDa, fractions 56–68). The smaller Sam68 complex was more prevalent in MCF-7 than BT20 cells (Fig. 1A). Another human breast adenocarcinoma epithelial cell line, MDA-MB-231, only harbored the smaller Sam68 complex (Fig. 1A). We also fractionated immortalized MEFs (Fig. 1A) and human normal diploid fibroblasts (not shown) and observed the presence of only the small Sam68 complex. These findings suggest that normal cells harbor Sam68 within the small complex and that certain transformed cells harbor Sam68 within a large or a large and a small complex.
FIGURE 1.
Sam68 resides within a large and a small complex. The small complex correlates with the alternative splicing activity of Sam68. A, cellular extracts prepared from HeLa, BT-20, MCF-7, MDA-MB-231, and MEFs were fractionated over a Superose 6 column to separate complexes according to size. The molecular mass markers for the gel filtration column are shown below. The proteins from each fraction (numbered from 35 to 80 shown above) were separated by SDS-PAGE, and the presence of Sam68 was detected by immunoblotting with anti-Sam68 antibodies. The migration of Sam68 is shown. B, HeLa, BT-20, MCF-7, MDA-MB-231, and MEFs were transfected with a CD44 minigene, and the level of inclusion of the V5 exon was assessed by reverse transcriptase-PCR. The 1-kb ladder molecular weight markers (Invitrogen) are shown on the left. The ratio of inclusion (+V5)/exclusion (−V5) was quantified by densitometric scanning and normalized to 1.00 using HeLa cells.
We next examined whether there was a correlation between the size of the Sam68 complex and the ability of Sam68 to alternatively splice the variable exon 5 (V5) of CD44, a known function of Sam68 (17). A CD44 minigene was transfected in the above-mentioned cell lines, and the level of V5 inclusion was assessed by RT-PCR (Fig. 1B; cd44 +V5). We observed that cell lines predominantly harboring the smaller Sam68 complex including MCF-7, MDA-MB-231, and MEFs displayed an elevated level (5–7-fold) of CD44 V5 exon inclusion compared with HeLa cells (Fig. 1B). BT-20 cells displayed a significant increase in exon V5 inclusion (2-fold) (Fig. 1B). These findings suggest that Sam68 exists in at least two molecular complexes and that the presence of the small Sam68 complex correlates with the ability of Sam68 to alternatively splice exon V5 of CD44.
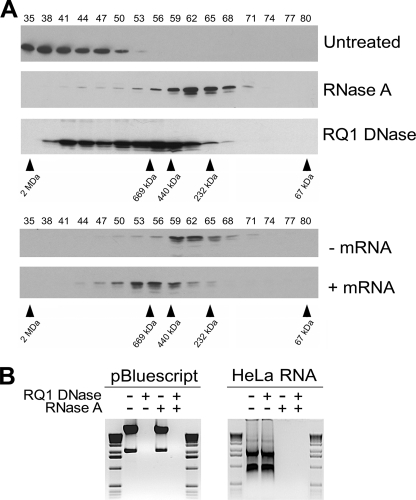
As Sam68 has been shown to associate with RNA and DNA (1), we treated HeLa cell lysates with RNase A or RNase-free DNase I and subsequently verified the mass of the Sam68 complex by gel filtration. The large complex of >1 MDa observed in HeLa cells was converted to the smaller complex with RNase A treatment (Fig. 2A, compare Untreated and RNase A), suggesting that the smaller complex is devoid of the majority of the RNA most likely associated with the Sam68 large complex. To further confirm that RNA accounts for the difference between the large and small Sam68 complexes, we isolated and converted the small complex into the larger complex with the addition of exogenous total RNA (Fig. 2A, lower panels). HeLa cell lysates preincubated with RNase A were separated on a Superose 6 column, and fractions 56–65 were collected, pooled, and concentrated using a Centricon YM-30. The resulting lysate was divided in two portions, one left untreated and the second incubated with total RNA purified from HeLa cells. Both samples were analyzed by gel filtration, and the addition of total RNA to the sample provoked an increase of the small complex (fractions 56–68) to a newly formed complex of 400–800 kDa of intermediate size (Fig. 2A, fractions 50–62). These data demonstrate that the addition of exogenous RNA partially converts the small Sam68 complex into a larger complex. Failure of complete restoration of the large complex could be due to lack of proper RNA folding, the absence of specific RNA targets, or other proteins dissociated during the process.
FIGURE 2.
Sam68 resides within an RNP complex in HeLa cells. A, HeLa cell lysates were left untreated or treated with RNase A and RNase-free DNase I (RQ1 DNase) and fractionated using a Superose 6 column. The presence of Sam68 was analyzed by immunoblotting. RNase A-treated fractions 56–66 were collected, pooled, and concentrated with using a Centricon YM-30. The lysates were divided in two; one was left untreated (−mRNA), and the second was incubated with HeLa total RNA (+mRNA). These cell lysates were subsequently fractionated over a Superose 6 column. B, the specificity of the RNase and DNase treatments was confirmed by incubating pBluescript DNA and total RNA isolated from HeLa cells with the indicated enzyme(s), and the products were separated on 1% agarose gels and visualized by ethidium bromide staining. The 1-kb ladder molecular weight markers (Invitrogen) are shown on the left and the right.
We also noted that DNase I treatment also decreased the size of the complex (fractions 38–65), although this decrease in complex size proved to be less dramatic than the one obtained with RNase A treatment (Fig. 2A, fractions 56–68). The partial reduction of the large Sam68 complex with DNase I treatment suggests that some Sam68 may be chromatin-associated. This is consistent with the ability of Sam68 to associate with DNA by chromatin immunoprecipitation assays (9, 21). The specificity of the RNase A and DNase I was confirmed using pBluescript and HeLa total RNA (Fig. 2B). Overall these findings suggest that the >1-MDa Sam68 complex is an RNP complex that may be chromatin-associated.
The Sam68 Complex Size Is Regulated by Growth Factors
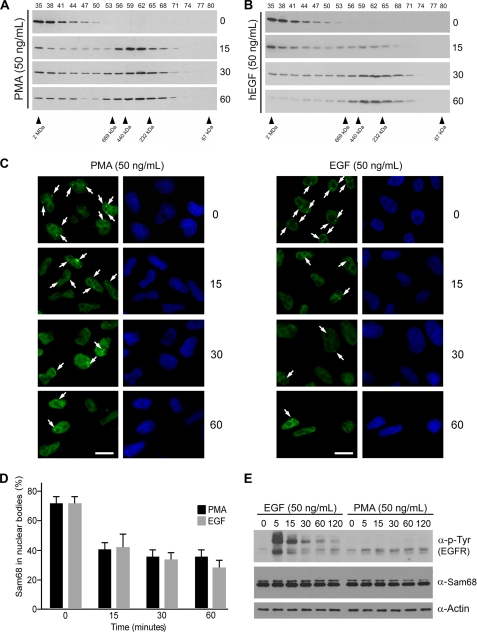
The addition of PMA induces the serine/threonine phosphorylation of Sam68 and promotes the inclusion of exon V5 of CD44 (17). In Fig. 1B we observed that the smaller Sam68 complex correlated with the alternative splicing activity of Sam68. Because PMA induces the alternative splicing activity of Sam68, we wished to link the two observations and examine if PMA treatment could alter the mass of the large >1 MDa Sam68 complex in HeLa cells. Indeed, PMA treatment promoted the appearance of the small Sam68 complex within 15 min and prolonged treatment of the 30- and 60-min established on equilibrium between the Sam68 complexes (Fig. 3A). These findings demonstrate that the large Sam68 complex is dynamic and can be converted into the smaller Sam68 complex with PMA. Another extracellular signal that results in the phosphorylation of Sam68 is EGF stimulation (36). The addition of 50 ng/ml EGF for 15 and 30 min also caused a progressive shift from the large complex to the smaller complex (Fig. 3B). At 60 min the large complex was almost completely converted to the smaller complex (Fig. 3B). These findings show that certain extracellular signals such as PMA and EGF may affect the alternatively splicing activity of Sam68 by modulating the mass of the Sam68 RNP complexes.
FIGURE 3.
EGF and PMA reduce the mass of the Sam68 RNP complex and diminish the presence of Sam68 in SNBs. A and B, HeLa cells stimulated with 50 ng/ml PMA or human EGF (hEGF) for 0, 15, 30, and 60 min were lysed, and the cell lysates were fractionated over a Superose 6 column, respectively. The presence of Sam68 in each fraction was assessed by immunoblotting with anti-Sam68 antibodies. C, HeLa cells treated with 50 ng/ml PMA or EGF were fixed in 1% paraformaldehyde, and the presence of Sam68 in nuclear bodies was assessed after 0-, 15-, 30-, and 60-min incubations by indirect immunofluorescence. D, the presence of Sam68 in nuclear bodies was carried out in 3 separate experiments as described in panel C in which 250–500 cells were counted for each time point. E, the Sam68 protein level was not affected by EGF or PMA stimulation. The level of activation of the EGF receptor (EGFR) was assessed using anti-phosphotyrosine (4G10) antibodies, and the level of Sam68 expression using Sam68-specific antibodies and β-actin antibodies was used to monitor equivalent expression.
The Presence of Sam68 in Nuclear Foci or SNBs Is Diminished with EGF Stimulation
The presence of the >1-MDa Sam68 complex correlates with the previously reported high prevalence of SNBs in HeLa cells (25). We wished to examine the ability of EGF and PMA treatments to regulate the dynamics of Sam68 within these foci. Using indirect immunofluorescence, we observed that the treatment of HeLa cells with PMA or EGF resulted in the rapid reduction of Sam68-positive foci as early as at 15 min of treatment (Fig. 3C and quantified in Fig. 3D). These observations suggest that the presence of Sam68 in nuclear foci correlates with Sam68 residing in the large >1-MDa complex and is modulated by EGF stimulation and PMA treatment. The level of tyrosine phosphorylation induced by EGF is shown in Fig. 3E, upper panel, and both EGF stimulation and PMA treatment did not reveal any change in the level of Sam68 protein expression (Fig. 3E, middle panel), suggesting that Sam68 is likely relocalized after stimulation and not the result of de novo protein synthesis.
Identification of the Protein Composition of the >1-MDa Sam68 Complex
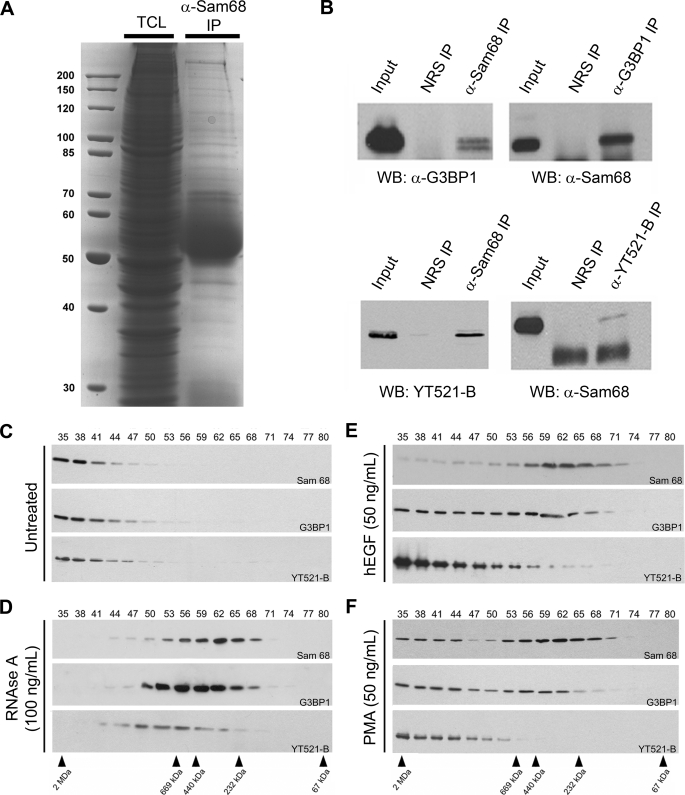
To identify the protein components constituting the large Sam68 complex, HeLa cells were fractionated, fractions 35–50 were pooled, and the Sam68 complex was immunopurified. The bound proteins (Fig. 4A) were excised from Coomassie Colloidal Blue-stained gel and identified using LC-MS/MS mass spectrometry (Table 1). We identified 44 proteins within the large Sam68 complex that had significant scores exceeding 43 that translates in a p value <0.05 (Table 1). Of these proteins, 45% were RNA binding and processing proteins implicated in various aspects of RNA metabolism, consistent with Sam68 residing within a large RNP complex. Another 41% of the identified proteins were associated with cell growth and cell cycle categories. The last 14% are part of pathways regulating cell adhesion and morphology. Of the 44 proteins that we identified, the interactions between Sam68 and hnRNP K, SLM-1, and SLM-2 have been previously reported (27, 37). We did not identify other known Sam68 interacting proteins such as Src kinases, the tyrosine kinase breast tumor kinase the splicing factor YT521-B, and others highlighted on the human Interactome website. However, ∼28% of the proteins, denoted with asterisks in Table 1, were identified by quantitative mass spectrometry in spreading initiation centers (SICs), structures that precede the formation of focal adhesion sites during cell adhesion (38).
FIGURE 4.
The RNA-binding protein G3BP1 is a component of the EGF-regulated Sam68 RNP complex. A, HeLa cell lysates were lysed, a fraction was kept to represent the total cellular lysate (TCL), and the remaining lysate was incubated with anti-Sam68 antibodies. The bound proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. The mass of the markers is shown on the left in kDa. IP, immunoprecipitate. B, shown is coimmunoprecipitation of endogenous Sam68 with G3BP1, a newly identified component of the Sam68 complex, and coimmunoprecipitation of Sam68 with YT521-B, a protein localized in SNBs that has previously been shown to associate with Sam68. Control immunoprecipitation was performed using normal rabbit serum (NRS). WB, Western blot. C and D, HeLa cell extracts were either left untreated or treated with 100 ng/ml RNase A. These cell extracts were fractionated over a Sepharose 6 column, and the fractions were immunoblotted with anti-Sam68, anti-G3BP1, and YT521-B antibodies as indicated. The fractionation of the protein markers is shown below in kDa. E and F, HeLa cells were stimulated with 50 ng/ml human EGF (hEGF) or PMA for 60 min, and cell extracts were prepared and fractionated as described in panels C and D.
TABLE 1.
Identification of the protein components of the Sam68 RNP complex
The protein components of Sam68 RNP complex are identified by LC-MS/MS mass spectrometry.
| Gene ID | Score | Protein name |
|---|---|---|
| Cancer, cell cycle, and proliferation | ||
| 10006 | 84 | Abl-interactor 1 |
| 4869 | 152 | B23 nucleophosmin |
| 8531 | 103 | Cold shock domain protein A |
| 1809 | 538 | Dihydropyrimidinase-like 3 |
| 10243 | 66 | Gephyrin |
| 3313 | 375 | Heat shock 70-kDa protein 98 (mortalin-2) |
| 3320 | 347 | Heat shock 90-kDa protein |
| 3065 | 71 | Histone deacetylase 1 |
| 9961 | 3.50E+05 | Major vault protein (Irp)a |
| 5340 | 45 | Plasminogen |
| 5154 | 50 | Platelet-derived growth factor receptor β precursor variant |
| 5315 | 370 | Pyruvate kinase (M2-PK)a |
| 6418 | 122 | SET translocation (myeloid leukemia-associated) |
| 7052 | 157 | Transglutaminase 2 isoform a |
| 7168 | 114 | Tropomyosin 1 |
| 7173 | 184 | Iropomyosin 3 |
| 7184 | 68 | Tumor rejection antigen (gp96) |
| 7415 | 145 | Valosin-containing protein |
| Transcription, splicing, and RNA metabolism | ||
| 1975 | 49 | elF4B |
| 10146 | 77 | G3BPIa |
| 3309 | 376 | Heat shock 70-kDa protein 5 (glucose-regulated protein, 78 kDa) |
| 3182 | 104 | hnRNP A/B |
| 3183 | 84 | hnRNP C |
| 3184 | 141 | hnRNP Da |
| 3190 | 87 | hnRNP Ka |
| 3191 | 65 | hnRNP L |
| 10236 | 87 | hnRNP R |
| 4691 | 97 | Nucleolin |
| 6202 | 244 | Ribosomal protein S8 |
| 3921 | 121 | Ribosomal protein SA (67 kDa laminin receptor)a |
| 6176 | 50 | Ribosomal protein, large, P1 |
| 6181 | 87 | Ribosomal protein, large, P2a |
| 6421 | 60 | SFPQ proteina |
| 202559 | 73 | SLM-1 |
| 10656 | 96 | SLM-2 |
| 6426 | 275 | Splicing factor; arginine/serine-rich 1 |
| 6427 | 141 | Splicing factor; arginine/serine-rich 2 |
| 10492 | 163 | Synaptotagmin binding; cytoplasmic RNA interacting protein |
| Cell adhesion and mobility | ||
| 58 | 184 | Actin αa |
| 60 | 206 | Actin βa |
| 70 | 127 | Actin γ |
| 2335 | 518 | Fibronectin 1a |
| 7431 | 118 | Vimentina |
a These proteins were also identified by quantitative mass spectrometry in SICs (38).
It is interesting to note that certain RNA-binding proteins, known to be substrates of tyrosine kinases, were identified. The tyrosine phosphorylation of hnRNP A/B is known to impair its RNA binding activity (39). hnRNP K has been identified to be an SH3-binding protein and a substrate of Src kinases (8). Ras-GAP-binding protein 1 (G3BP1) is an RNA-binding protein with signaling properties that can bind SH3 domains and is a substrate of tyrosine kinases (40, 41). We proceeded with the validation of the G3BP1/Sam68 association and examined the known interaction between Sam68/YT521-B, a known tyrosine-phosphorylated protein that localizes to SNBs (28). The association between G3BP1/Sam68 and Sam68/YT521-B was observed by reciprocal immunoprecipitations using endogenous proteins of HeLa cell extracts (Fig. 4B). To verify whether G3BP1 and YT521-B are part of the large Sam68 complex, we immunoblotted the fractions prepared from HeLa cell extracts with anti-G3BP1 and anti-YT521-B antibodies. Indeed both G3BP1 and YT521-B co-purified in the large Sam68 complex (Fig. 4C). G3BP1 was observed to migrate with Sam68 in the small complex when cells were treated with RNase A, PMA, or EGF (Fig. 4, D–F). In contrast, treatment with either PMA or EGF did not affect the presence of YT521-B within the small complex; however, YT521-B did localize within the small Sam68 complex with RNase A treatment (Fig. 4, E and F). The EGF-induced small complex was immunopurified and visualized by Coomassie staining to determine whether the small complex likely represents new protein components or is likely a breakdown of the large complex. The mass of the Sam68-associated proteins was similar with or without EGF treatment (supplemental Fig. 1), suggesting that the small complex is likely a subset of the larger complex.
To assess the role of phosphorylation of Sam68 in the conversion of the large complex to the small complex, we monitored the phosphorylation of Sam68 tyrosine 440, a site known to be phosphorylated in an EGF-induced manner (36). Sam68 was immunoprecipitated with anti-Sam68 antibodies from BT-20, HeLa, MCF-7, and MDA-MB-231 and MEFs, and the level of phosphorylation of Tyr-440 examined using a site-specific antibody (36). We detected similar levels of phosphorylation at Tyr-440 in all cell lines tested regardless of the Sam68 complex size it harbors (supplemental Fig. 2). These findings suggest that phosphorylation of Sam68 can occur in both complexes.
Growth Factors Promote the Inclusion of Exon V5 of CD44
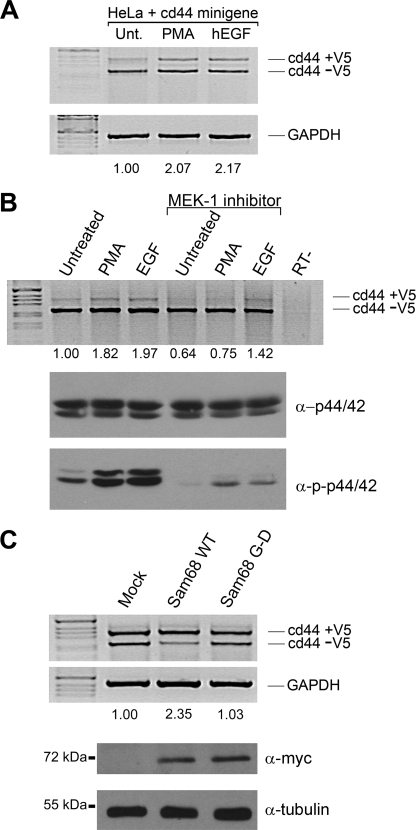
We next examined the ability of EGF to promote the inclusion of CD44 V5 exon and whether the MAP kinase pathway is required. HeLa cells were transfected with the CD44 V5 reporter gene, and the cells were left untreated (negative control) or treated with either PMA (positive control) or EGF. We observed a significant 2-fold increase in the inclusion of the V5 exon in the presence of PMA or EGF stimulation (Fig. 5A). RT-PCR for glyceraldehyde-3-phosphate dehydrogenase was used to confirm equivalent amounts of RNA were utilized. We next tested whether the MEK-1 inhibitor (U0126) was able to inhibit the PMA-induced inclusion of V5. The PMA-induced V5 inclusion was completely blocked with U0126 (Fig. 5B), as reported previously (17). In contrast, the EGF-induced inclusion of CD44 V5 exon was only partially abrogated by the MEK-1 inhibitor (Fig. 5B), suggesting that EGF induces V5 inclusion by MAP kinase-dependent and -independent pathways. The confirmation that U0126 treatment worked was demonstrated using anti-phosphospecific p44/42 antibodies normalized with anti-p44/42 antibodies (Fig. 5B).
FIGURE 5.
EGF regulates Sam68 dependent splicing of CD44 exon V5 and is partially dependent on the activation of the MAP kinase pathway. A, HeLa cells transfected with a CD44 minigene reporter plasmid were left untreated (Unt) or stimulated with 50 ng/ml PMA or human EGF (hEGF) for 3 h. Total RNA was isolated, and the inclusion of exon V5 was assessed by RT-PCR followed by separation of the PCR-generated DNA fragments by agarose gel electrophoresis. The percentage of inclusion of V5 was determined by densitometric scanning and was normalized to the untreated cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized to confirm equivalent RNA was present in each sample. B, the MEK inhibitor (U0126) was added (100 nm) in the presence of 50 ng/ml PMA or EGF for 3 h. The level of activation of p44/42 was determined with the phosphospecific antibody directed against phospho-p44/42 and compared with total p44/42. C, Sam68 null MEFs were transfected with a CD44 minigene reporter plasmid in the presence of an expression vector encoding Myc-Sam68 or Myc-Sam68 G178D, an amino acid substitution known to abrogate RNA binding. The inclusion of exon V5 was assessed RT-PCR. The expression of the Myc epitope-tagged Sam68 proteins was confirmed by immunoblotting. Anti-β-tubulin antibodies were used to confirm equivalent protein expression. WT, wild type.
To assess if the Sam68 RNA binding domain was essential for the V5 inclusion, we transfected Sam68 null MEFs with expression vectors encoding wild-type Sam68 or RNA binding-defective protein (Sam68: G178D) (42). We utilized Sam68 null MEFs to avoid the background of the endogenous Sam68 protein. Wild-type Sam68, but not the Sam68·G178D RNA binding-defective protein, was able to increase CD44 V5 exon inclusion (Fig. 5C; compare 2.35- versus 1.03-fold). The expression of the Myc epitope-tagged Sam68 protein was equivalent, as assessed by immunoblotting (Fig. 5C). These findings suggest that Sam68, through its RNA binding domain, can facilitate the inclusion of CD44 V5 and that this activity is regulated by both PMA and EGF stimulation.
Sam68-deficient Cells Have Impaired Cell Migration
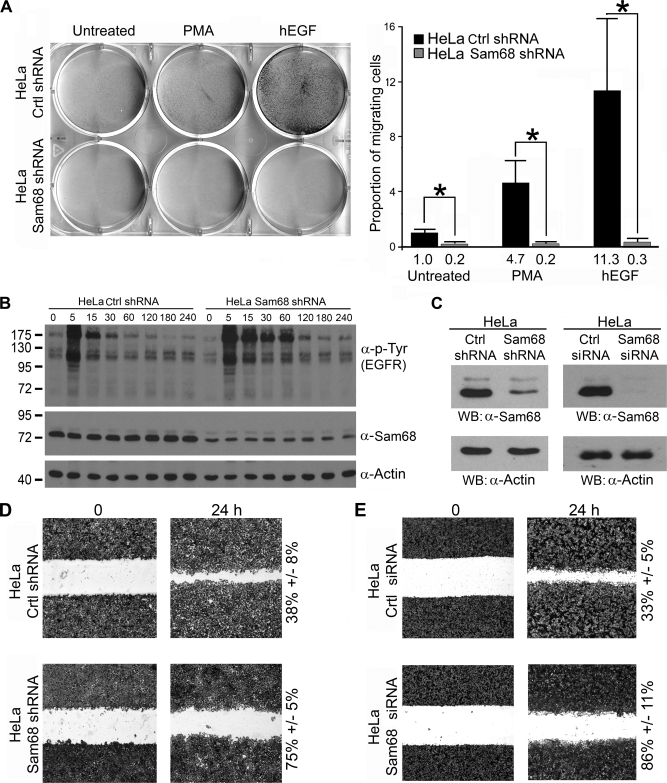
We have demonstrated that EGF, like PMA, promotes the inclusion of CD44 V5 exon (Fig. 5A). The expression of CD44 variants correlates with invasiveness of certain cancers (18, 34). This prompted us to investigate whether Sam68 regulated PMA- and EGF-induced chemotactic cell migration. We reduced the expression of Sam68 in HeLa cells using a stably integrated pSUPER-based vector harboring a Sam68sh sequence. The expression of Sam68 was reduced >70% in Sam68sh cells compared with pSUPER controls (Fig. 6, B and C) and >95% using an siGENOME siRNA that targets Sam68 (Fig. 6C). We observed that the Sam68-deficient cells had decreased migration toward EGF compared with control HeLa cells (Fig. 6A). This was not due to a lack of EGF receptor activation, as the EGF receptor showed increased activation and phosphorylation with exposure time to EGF (Fig. 6B, upper panel). We noted the presence of elevated tyrosine phosphorylation in Sam68-deficient HeLa cells, and this is consistent with our previous observations in Sam68 null cells (3, 43).
FIGURE 6.
Sam68-deficient HeLa cells have impaired EGF-induced cell migration. A, HeLa cells stably transfected with pSUPER (Ctrl shRNA) or Sam68 shRNA were examined for their ability to migrate toward PMA and human EGF (hEGF) using a Transwell migration assay. The cells that migrated are shown using crystal violet staining. The quantification is indicated on the right, and the migrating cells were normalized to the number of untreated HeLa Ctrl shRNA (counted in four random experiments) that migrated to the lower chamber. (*, p value ≤0.005) B, the expression of Sam68, β-actin, and the tyrosine phosphorylation of the EGF receptor (EGFR) in pSUPER control and Sam68sh HeLa cells was assessed by immunoblotting with anti-Sam68, anti-β-actin, and anti-phosphotyrosine (4G10) antibodies. C, shown is the Sam68 expression level in HeLa Sam68 shRNA cells versus pSuper control cells. In addition, HeLa cells were treated with an siGENOME siRNA that targets Sam68 or a control siRNA. The level of Sam68 expression was assessed by immunoblotting (WB) with anti-Sam68 and compared with the level of β-actin. D, pSUPER and Sam68sh HeLa cells were plated on fibronectin-coated tissue culture dishes, and a wound was introduced. The directional migration of the cells was visualized by crystal violet staining. E, HeLa cells were transfected with siGENOME Sam68 siRNA or control small interfering GFP. The directional migration of the cells was visualized by crystal violet staining.
PMA was a less potent chemoattractant compared with EGF; nevertheless, HeLa cells harboring Sam68 shRNA were also impaired for PMA-induced cell migration (Fig. 6A). These findings demonstrate that Sam68 is required for growth factor-induced cell migration. To assess the directional migration capabilities of Sam68-deficient HeLa cells, a wound-healing assay was performed. As expected, decreased expression of Sam68 with either shRNA or siRNA correlated with decreased directional migration (compare 75% for Sam68sh versus 38% for control and 86% for the siSam68 versus 33% for the control) (Fig. 6, D and E). These findings show that Sam68 is required for EGF- and PMA-induced cell migration.
DISCUSSION
In the present study we identify a Sam68 complex with a mass >1 MDa in HeLa cells that is composed of ∼40 proteins using an immunoprecipitation/mass spectrometry approach. We observed that the existence of this complex correlates with the presence of Sam68 in nuclear foci termed Sam68 nuclear bodies. The large Sam68 complex and the presence of Sam68 in nuclear bodies was regulated by EGF, as it reduced the size of the Sam68 complex to a mass of ∼200–450 kDa and diminished the presence of Sam68 in nuclear foci. In addition to its role in cell migration, we showed that EGF induced the alternative splicing of CD44 variable exon V5, events that were Sam68-dependent. Our findings suggest that in transformed cells such as HeLa cells, Sam68 exists in a >1-MDa RNP complex that is regulated by growth factor signaling.
RNP complexes consist of many specific RNA-binding proteins and RNAs. These RNPs are involved in the regulation of essentially all aspects of RNA metabolism (44). RNPs are often localized in macromolecular structures termed RNA granules or more specialized structures such as stress granules and transport granules, and they share common proteins with similar functions (45, 46). As examples, the fragile X mental retardation protein and Staufen RNA-binding proteins are known to be part of large RNP complexes with specific mRNAs (47, 48). The fragile X mental retardation protein and Staufen RNPs are stored in RNA granules that consist mainly of repressed messenger RNAs (49–52). In our present study many protein components of the Sam68 RNP complex that we identified are involved in mRNA processing (Table 1). Our proteomic analysis of the Sam68 RNP complex permitted us to identify ∼40 proteins. Surprisingly, we fail to identify known interactors of Sam68 such as YT521-B and breast tumor kinase in our proteomic studies. This is probably because of their low level of expression and their low affinity for Sam68. On the other hand, we identified G3BP1 as part of the Sam68 complex. Although predominantly cytoplasmic, G3BP1 has the capability to enter the nucleus (41, 53, 54). It is still unknown if Sam68 mediated this relocalization of a small proportion of G3BP1 to the nucleus through the formation of the RNP complex. From the ∼40 proteins identified, ∼28% of these proteins are common, with the protein components of RNP complexes localized in a membrane structure termed the SIC (38). We also noted that some of the proteins from the Sam68 RNPs are also constituents of RNA granules. These proteins include eIF4B, G3BP, hsp70, nucleolin, syncrip, SFPQ (splicing factor proline/glutamine-rich), hnRNPs, and ribosomal components (45, 50, 54, 55). These findings suggest that the components of the Sam68 RNP complex partially overlap with the components of repressed RNPs observed in RNA granules, indicating a role for Sam68 in translational control of specific mRNAs. Indeed, a recent study shows that a cytoplasmic Sam68 mutant can sequester HIV negative factor mRNA in stress granules, thus drastically decreasing the translation of negative factor (16). Moreover, we showed that Sam68 is directly implicated in the polysomal recruitment of specific mRNAs during spermatogenesis (13). The Sam68 RNPs may also regulate nuclear events such as mRNA splicing, as Sam68 was shown to regulate the splicing of specific mRNAs during neurogenesis (20). Because Sam68 is localized in structures such as SICs and SNBs, RNP storing structures responsive to extracellular stimuli, the presence of Sam68 could enable mobilization of certain RNPs.
Our data demonstrate that growth factors can regulate the mass of the Sam68 complex in transformed cells and regulate the localization of Sam68 within nuclear bodies. These observations also correlate with the increased migration observed with EGF and PMA stimulation. How could growth factor-induced cellular migration be impaired with the loss of Sam68? Implication of RNA-binding proteins in cellular migration and adhesion was previously shown for RNA-binding proteins present in SICs (38). De Hoog et al. (38) showed that the electroporation of antibodies against the RNA-binding proteins hnRNP E1, hnRNP K, and TLS/FUS (translocated in liposarcoma) blocked cell migration. More recently, it was shown that hnRNP K (56), G3BP1 (41), and Sam68 regulate cell migration (3). In the latter case, we show that Sam68 functions as an adaptor protein during integrin signaling (3). It is likely that during cell migration Sam68 plays a dual role as an RNA-binding protein and as an adaptor protein depending on the stimuli. It was previously shown that RNA-binding proteins involved in cell migration such as hnRNP K and G3BP1 are substrates of tyrosine kinases (41, 57), similar to Sam68 (1), which is consistent with signaling cascades regulating the dynamics of these complexes.
The change in mass of the Sam68 complex after EGF and PMA treatment in HeLa cells correlated with the splicing activities of Sam68 (2-fold), albeit not as much as observed in certain cell lines such as MCF-7 (7-fold) cells that only harbor the small complex. The alternative splicing in MCF-7 cells took place during a 48-h time frame, whereas the alternative splicing was assessed 3 h after PMA and EGF stimulation. Cell-specific factors may also be a major contribution to the observed difference. These findings also suggest that the formation of the Sam68 large complex may restrict Sam68 to these structures such as with SNBs. Even if the mechanism regulating the equilibrium between the large and small complexes remains unknown, it is clear that post-translational modifications of Sam68 play a role in Sam68 RNA binding activity and its cellular localization (25, 42). Because PMA can also promote the formation of the small complex (Fig. 3A) without the activation of the EGF receptor (Fig. 3E), comparison of the effect of EGF and PMA in a functional model will likely unveil the in-depth mechanism behind the modulation of Sam68 complex size, its influence in splicing activity, and how this affects cellular migration.
Immunoprecipitation of the Sam68 complex after EGF treatment resulted in the enrichment of essentially the same proteins as in the large complex, albeit with a slight reduction (supplemental Fig. 1). This suggests that the smaller complex may be the result of the breakdown of the larger complex. Growth factor signaling has the ability to activate multiple cis- and trans-acting factors that can target RNA-binding proteins and affect their affinity for mRNA targets. The phosphorylation of serine/arginine-rich proteins, for example, is necessary to render serine/arginine-rich proteins specific RNA-binding proteins (58). This seems to be the case with Sam68, where growth factor stimulation was shown to induce its phosphorylation (36). These events stimulate the Sam68 alternative splicing activity and increase the inclusion of specific exons such as exon 5 of CD44 (17). In addition, regulation of alternative splicing of the Bcl-x gene by Sam68 is tightly regulated by tyrosine phosphorylation by Src family tyrosine kinase Fyn (19).
The correlation between EGF receptor amplification and cancer is well documented (32, 33, 35). Sam68 is tyrosine-phosphorylated in breast tumor cell lines and tissues (36, 59), and its coexpression with the breast tumor kinase tyrosine kinases in tumors can serve as a prognostic marker for long time survival (60). Moreover, the expression of the Sam68-associated protein, G3BP1, is observed in >80% of epithelial cancers including breast tumors, and its expression correlates with lymph node metastasis and c-erbB2 overexpression (61). These findings suggest that the phosphorylation of Sam68 and its associated proteins in the >1-MDa Sam68 RNP complex including hnRNP A/B, hnRNP K, and G3BP1 by growth factor signaling pathways can regulate cancer metastasis and invasiveness. Therefore, the phosphorylation of Sam68 RNP complex components may be markers for growth factor-induced signaling. Inhibitors of Sam68 function or compounds that disrupt Sam68 RNP complexes may be useful to block cell metastasis and invasiveness.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Grant MT13377.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- Sam68
- Src associated in mitosis of 68 kDa
- SH
- Src homology
- siRNA
- small interfering RNA
- RNP
- ribonucleoprotein
- hnRNP
- heterogeneous nuclear RNP
- EGF
- epidermal growth factor
- SNB
- Sam68 nuclear body
- MEF
- mouse embryo fibroblast
- PMA
- phorbol 12-myristate 13-acetate
- SIC
- spreading initiation center
- G3BP1
- Ras-GAP-binding protein 1
- HIV
- human immunodeficiency virus
- SLM
- Sam68-like mammalian family members
- MAP
- mitogen-activated protein
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- RT
- reverse transcription
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Lukong K. E., Richard S. (2003) Biochim. Biophys. Acta. 1653, 73–86 [DOI] [PubMed] [Google Scholar]
- 2.Fumagalli S., Totty N. F., Hsuan J. J., Courtneidge S. A. (1994) Nature 368, 871–874 [DOI] [PubMed] [Google Scholar]
- 3.Huot M. E., Brown C. M., Lamarche-Vane N., Richard S. (2009) Mol. Cell. Biol. 29, 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maa M. C., Leu T. H., Trandel B. J., Chang J. H., Parsons S. J. (1994) Mol. Cell. Biol. 14, 5466–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard S., Yu D., Blumer K. J., Hausladen D., Olszowy M. W., Connelly P. A., Shaw A. S. (1995) Mol. Cell. Biol. 15, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor S. J., Shalloway D. (1994) Nature 368, 867–871 [DOI] [PubMed] [Google Scholar]
- 7.Vogel L. B., Fujita D. J. (1995) J. Biol. Chem. 270, 2506–2511 [DOI] [PubMed] [Google Scholar]
- 8.Weng Z., Thomas S. M., Rickles R. J., Taylor J. A., Brauer A. W., Seidel-Dugan C., Michael W. M., Dreyfuss G., Brugge J. S. (1994) Mol. Cell. Biol. 14, 4509–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung N., Chan L. C., Thompson A., Cleary M. L., So C. W. (2007) Nat. Cell Biol. 9, 1208–1215 [DOI] [PubMed] [Google Scholar]
- 10.Volk T., Israeli D., Nir R., Toledano-Katchalski H. (2008) Trends Genet. 24, 94–101 [DOI] [PubMed] [Google Scholar]
- 11.Lukong K. E., Chang K. W., Khandjian E. W., Richard S. (2008) Trends Genet. 24, 416–425 [DOI] [PubMed] [Google Scholar]
- 12.Lin Q., Taylor S. J., Shalloway D. (1997) J. Biol. Chem. 272, 27274–27280 [DOI] [PubMed] [Google Scholar]
- 13.Paronetto M. P., Messina V., Bianchi E., Barchi M., Vogel G., Moretti C., Palombi F., Stefanini M., Geremia R., Richard S., Sette C. (2009) J. Cell Biol. 185, 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy T. R., Xu W., Mau J. K., Goodwin C. D., Suhasini M., Tang H., Frimpong K., Rose D. W., Wong-Staal F. (1999) Nat. Med. 5, 635–642 [DOI] [PubMed] [Google Scholar]
- 15.Li J., Liu Y., Kim B. O., He J. J. (2002) J. Virol. 76, 8374–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henao-Mejia J., Liu Y., Park I. W., Zhang J., Sanford J., He J. J. (2009) Mol. Cell 33, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matter N., Herrlich P., König H. (2002) Nature 420, 691–695 [DOI] [PubMed] [Google Scholar]
- 18.Cheng C., Sharp P. A. (2006) Mol. Cell. Biol. 26, 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paronetto M. P., Achsel T., Massiello A., Chalfant C. E., Sette C. (2007) J. Cell Biol. 176, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chawla G., Lin C. H., Han A., Shiue L., Ares M., Jr., Black D. L. (2009) Mol. Cell. Biol. 29, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batsché E., Yaniv M., Muchardt C. (2006) Nat. Struct. Mol. Biol. 13, 22–29 [DOI] [PubMed] [Google Scholar]
- 22.Babic I., Cherry E., Fujita D. J. (2006) Oncogene 25, 4955–4964 [DOI] [PubMed] [Google Scholar]
- 23.Hong W., Resnick R. J., Rakowski C., Shalloway D., Taylor S. J., Blobel G. A. (2002) Mol. Cancer Res. 1, 48–55 [PubMed] [Google Scholar]
- 24.Richard S., Torabi N., Franco G. V., Tremblay G. A., Chen T., Vogel G., Morel M., Cléroux P., Forget-Richard A., Komarova S., Tremblay M. L., Li W., Li A., Gao Y. J., Henderson J. E. (2005) PLoS Genet. 1, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T., Boisvert F. M., Bazett-Jones D. P., Richard S. (1999) Mol. Biol. Cell 10, 3015–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denegri M., Chiodi I., Corioni M., Cobianchi F., Riva S., Biamonti G. (2001) Mol. Biol. Cell 12, 3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Fruscio M., Chen T., Richard S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2710–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann A. M., Nayler O., Schwaiger F. W., Obermeier A., Stamm S. (1999) Mol. Biol. Cell 10, 3909–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seales E. C., Shaikh F. M., Woodard-Grice A. V., Aggarwal P., McBrayer A. C., Hennessy K. M., Bellis S. L. (2005) J. Biol. Chem. 280, 37610–37615 [DOI] [PubMed] [Google Scholar]
- 30.Wu J., Dent P., Jelinek T., Wolfman A., Weber M. J., Sturgill T. W. (1993) Science 262, 1065–1069 [DOI] [PubMed] [Google Scholar]
- 31.Zheng C. F., Ohmichi M., Saltiel A. R., Guan K. L. (1994) Biochemistry 33, 5595–5599 [DOI] [PubMed] [Google Scholar]
- 32.Citri A., Yarden Y. (2006) Nat. Rev. Mol. Cell. Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 33.Nicholson R. I., Gee J. M., Harper M. E. (2001) Eur. J. Cancer 37, Suppl. 4, S9–15 [DOI] [PubMed] [Google Scholar]
- 34.Ponta H., Sherman L., Herrlich P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 35.Slamon D. J., Godolphin W., Jones L. A., Holt J. A., Wong S. G., Keith D. E., Levin W. J., Stuart S. G., Udove J., Ullrich A. (1989) Science 244, 707–712 [DOI] [PubMed] [Google Scholar]
- 36.Lukong K. E., Larocque D., Tyner A. L., Richard S. (2005) J. Biol. Chem. 280, 38639–38647 [DOI] [PubMed] [Google Scholar]
- 37.Yang J. P., Reddy T. R., Truong K. T., Suhasini M., Wong-Staal F. (2002) Oncogene 21, 7187–7194 [DOI] [PubMed] [Google Scholar]
- 38.de Hoog C. L., Foster L. J., Mann M. (2004) Cell 117, 649–662 [DOI] [PubMed] [Google Scholar]
- 39.Pype S., Slegers H., Moens L., Merlevede W., Goris J. (1994) J. Biol. Chem. 269, 31457–31465 [PubMed] [Google Scholar]
- 40.Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B. (1996) Mol. Cell. Biol. 16, 2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahmouni S., Vang T., Alonso A., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S. P., Mustelin T. (2005) Mol. Cell. Biol. 25, 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T., Damaj B. B., Herrera C., Lasko P., Richard S. (1997) Mol. Cell. Biol. 17, 5707–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richard S., Vogel G., Huot M. E., Guo T., Muller W. J., Lukong K. E. (2008) Oncogene 27, 548–556 [DOI] [PubMed] [Google Scholar]
- 44.Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. (2008) FEBS Lett. 582, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson P., Kedersha N. (2006) J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson P., Kedersha N. (2008) Trends Biochem. Sci. 33, 141–150 [DOI] [PubMed] [Google Scholar]
- 47.Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 48.Furic L., Maher-Laporte M., DesGroseillers L. (2008) RNA 14, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antar L. N., Dictenberg J. B., Plociniak M., Afroz R., Bassell G. J. (2005) Genes Brain Behav. 4, 350–359 [DOI] [PubMed] [Google Scholar]
- 50.Kanai Y., Dohmae N., Hirokawa N. (2004) Neuron. 43, 513–525 [DOI] [PubMed] [Google Scholar]
- 51.Mazroui R., Huot M. E., Tremblay S., Filion C., Labelle Y., Khandjian E. W. (2002) Hum. Mol. Genet. 11, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 52.Miki T., Takano K., Yoneda Y. (2005) Cell Struct. Funct. 30, 51–56 [DOI] [PubMed] [Google Scholar]
- 53.French J., Stirling R., Walsh M., Kennedy H. D. (2002) Histochem. J. 34, 223–231 [DOI] [PubMed] [Google Scholar]
- 54.Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003) J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Mokas S., Mills J. R., Garreau C., Fournier M. J., Robert F., Arya P., Kaufman R. J., Pelletier J., Mazroui R. (2009) Mol. Biol. Cell 20, 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue A., Sawata S. Y., Taira K., Wadhwa R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrowski J., Schullery D. S., Denisenko O. N., Higaki Y., Watts J., Aebersold R., Stempka L., Gschwendt M., Bomsztyk K. (2000) J. Biol. Chem. 275, 3619–3628 [DOI] [PubMed] [Google Scholar]
- 58.Misteli T., Cáceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F., Spector D. L. (1998) J. Cell Biol. 143, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babic I., Jakymiw A., Fujita D. J. (2004) Oncogene. 23, 3781–3789 [DOI] [PubMed] [Google Scholar]
- 60.Aubele M., Walch A. K., Ludyga N., Braselmann H., Atkinson M. J., Luber B., Auer G., Tapio S., Cooke T., Bartlett J. M. (2008) Br. J. Cancer 99, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ning J. Y., You J. F., Pei F., Wang J. L., Cui X. L., Zheng J. (2005) Zhonghua Bing Li Xue. Za. Zhi. 34, 215–219 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.