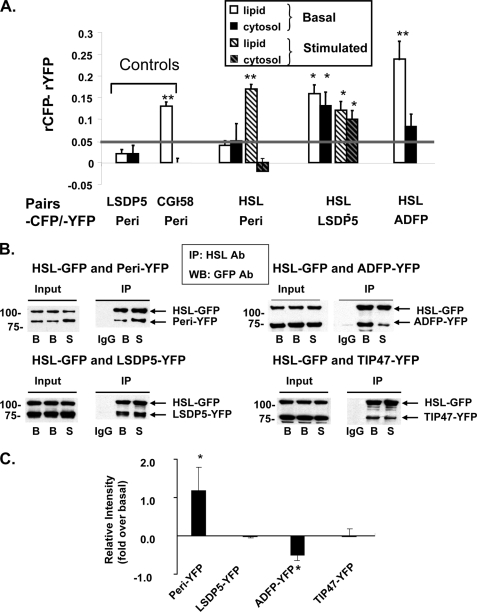
FIGURE 3.
HSL binds to PAT family proteins, as determined by in situ AFRET and co-immunoprecipitation (IP). A, images were collected by confocal microscopy of live cells expressing pairs of fluorescent fusion proteins shown along the x axis and calculations for AFRET were performed (29). The gray line indicates the threshold of significance. Data are means ± S.E. from 6 to 12 experiments. *, p < 0.05; **, p < 0.01. B, CHO-K1 cells expressing HSL-CFP were transfected with a PAT fusion protein as follows: perilipin-YFP, ADFP-YFP, or LDSP-5-YFP. The cells were then incubated with 400 μm oleic acid overnight. The following day, cells were incubated with 5 μm triacsin C (B, basal conditions) or with 10 μm forskolin, 1 mm IBMX, and 5 μm triacsin C for 30 min (S, stimulated conditions). Cell lysates were incubated with rabbit anti-HSL IgG (basal or stimulated) or rabbit pre-immune control IgG. Immunoprecipitates were analyzed by Western blot for PAT family proteins and HSL using a commercial GFP antibody that cross-reacts with YFP. One of three similar experiments is shown. IP, immunoprecipitation; WB, Western blot; Ab, antibody. C, Western blot ECL signals were quantified by densitometry using ImageJ software. Each data point represents the average of the calculated ratio of the amount of each PAT protein recovered in stimulated conditions relative to the amount recovered in basal conditions. Data represent means ± S.E. of three experiments (*, p < 0.05, basal versus stimulated conditions).