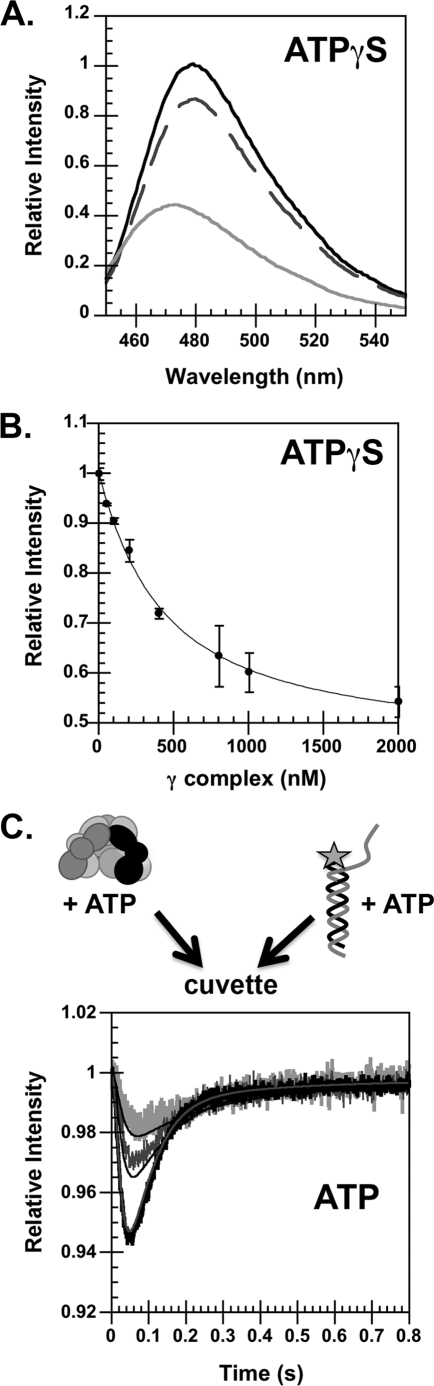
FIGURE 4.
Binding of γ complex to DNA. A, emission spectra of DCC were measured using an excitation wavelength of 455 nm. The black trace is a scan of free p/t-DNA-DCC, the dashed dark gray trace is a scan repeated after the addition of γ complex, and the light gray trace is a scan repeated after the addition of ATPγS. Final concentrations were: 100 nm p/t-DNA-DCC, 2000 nm γ complex, and 0.5 mm ATPγS in assay buffer. B, the relative intensity of DCC in a clamp loader·DNA complex was determined by measuring DCC fluorescence at 480 nm as a function of γ complex concentration. The maximal quench in DCC fluorescence was calculated by fitting these data to a quadratic equation (see “Experimental Procedures”). The solid line through the data is the result of the fit, which gave a value of 0.58 for the relative intensity of DCC in a clamp loader·DNA complex and a KD value of 375 ± 7 nm for clamp loader·DNA dissociation. Final concentrations were: 100 nm p/t-DNA-DCC and 0.5 mm ATPγS in assay buffer. C, the rate of γ complex binding to DNA was measured in reactions in which a solution of γ complex and ATP was added to a solution of p/t-DNA-DCC and ATP. The relative fluorescence of DCC is plotted as a function of time. Final concentrations were: 125, 250, and 500 nm γ complex and p/t-DNA-DCC and 0.5 mm ATP in assay buffer with 4% glycerol. The concentrations of γ complex and DNA are indicated by the shade of gray; darker indicates a higher concentration. Solid lines through the reaction time course were generated from the kinetic model illustrated in Fig. 8.