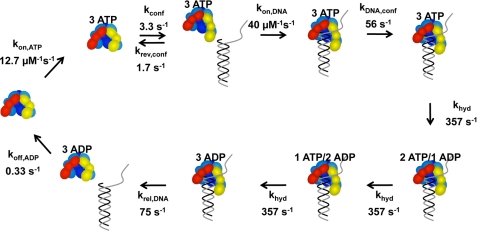
FIGURE 8.
Kinetic model fit to p/t-DNA binding data. The model starts on the left-hand side with free γ complex. (Note: the γ complex used in our studies contains seven polypeptides, γ3δδ′χψ, but only five, γ3δδ′, are illustrated in the diagram.) The γ complex binds 3 molecules of ATP in a single step. ATP binding promotes a conformational change that allows the clamp loader to bind DNA. DNA binding induces a second conformational change that activates the ATP sites for hydrolysis and three molecules of ATP are hydrolyzed sequentially at the same rate. DNA and then ADP are released to allow the γ complex to recycle. The relative intensity of free DNA was set at 1 and the relative intensities for all bound DNA species set at 0.58. The data in Figs. 4C, 6, and 7 were fit to this model using DynaFit (34).