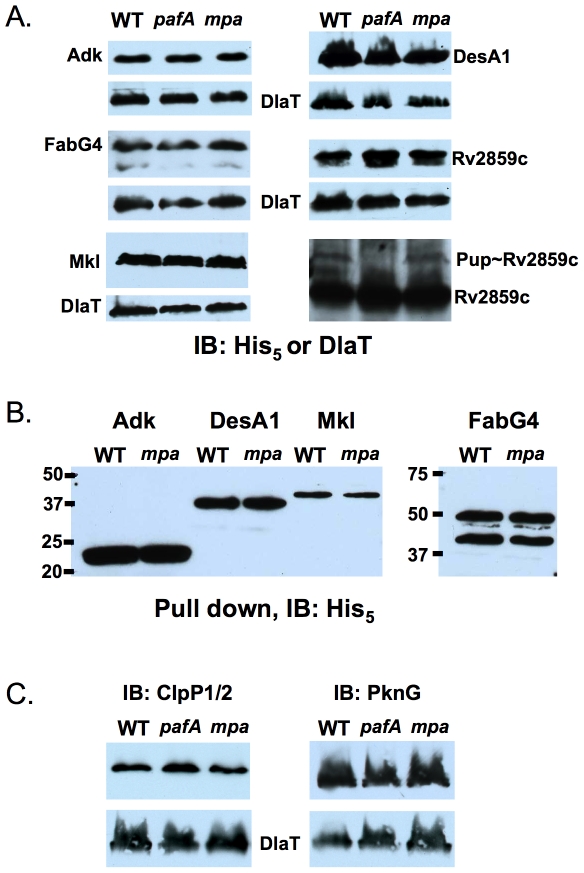
Figure 2. Steady state levels of several RCC pupylome proteins in wild type, pafA and mpa mutants.
(A) Five pupylation substrates do not show increased steady state levels in proteasome-defective Mtb. Adk, FabG4, Mkl, DesA1 and Rv2859c were all epitope tagged with His6 at the C-termini. FabG4-His6 is predicted to have a MW of 48 kD, corresponding with the top band; a lower MW species appears in all strains tested, perhaps indicating proteolytic processing or an alternative translation initiation site after the predicted start codon. DlaT loading control is shown below each anti-His5 immunoblot (IB). (B) Purification of His6-tagged Adk, DesA1, Mkl, and FabG4 from wild type and mpa Mtb strains did not identify pupylated proteins. The upper band is the presumed full length FabG4 (without Pup). (C) IB analysis shows that endogenous ClpP1/2 and PknG do not accumulate in proteasome defective Mtb under routine culture conditions. Equivalent cell numbers from cultures grown to an optical density (ODA580) = 1.5 were analyzed. DlaT is the loading control for all samples.