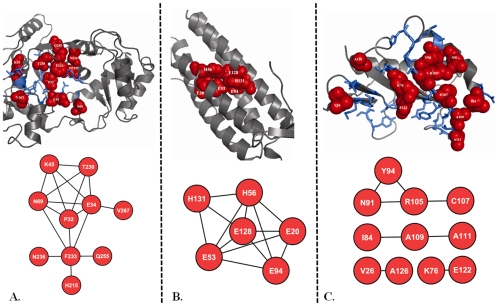
Figure 3. Examples of coevolved functional sites.
Coevolved (panel A, marked in red spheres) and non-coevolved (panel A, marked in blue sticks) active sites are projected onto the structure of a representative member (PDB code: 1EUQ) from Glutamyl-tRNA synthetase(GluRS)/Glutaminyl-tRNA synthetase (GlnRS) catalytic domain family (CDD code: CD00418). Here an edge connects two coevolved residues (red circles). Coevolved metal binding sites (panel B, marked in red spheres) are projected onto the 3D structure of a representative member (PDB code: 1LKO) from ferritin-like diiron-carboxylate protein domain family (CDD code: CD00657). Coevolved (panel C, marked in red spheres) and non-coevolved (panel C, marked in blue sticks) protein binding sites are projected onto the structure of a representative member (PDB code: 1JD1) from YjgF_YER057c_UK114_family (CDD code: CD00448). Protein structural and network representations were created using the PyMol [47] and Cytoscape [48] program.