Abstract
OBJECTIVE
Previous studies have indicated that the latent autoimmune diabetes in adults (LADA) phenotype is heterogeneous and that LADA patients share features of type 1 and type 2 diabetes in various proportions. We tested for association of known type 1 and type 2 diabetes susceptibility genes in LADA subjects and analyzed relationships to a marker of autoimmune activity (titers of anti-GAD) and a phenotypic risk factor of type 2 diabetes (BMI).
RESEARCH DESIGN AND METHODS
Data were assembled from the Nord-Trøndelag Health Study (HUNT) study, which comprises the adult population of an entire county in Norway. We genotyped 60 single nucleotide polymorphisms (SNPs) known to be associated with type 1 or type 2 diabetes, including 14 tag SNPs used for HLA haplotyping in 120 type 1 diabetic, 126 LADA, and 1,090 type 2 diabetic patients and 1,503 age- and sex-matched nondiabetic subjects.
RESULTS
The majority of the strongly associated HLA haplotypes for type 1 diabetes were significantly associated with LADA in general, but mainly with high anti-GAD LADA patients. Two distinct HLA haplotypes were associated only with LADA and mainly in low anti-GAD LADA patients. There were no associations of non-HLA type 1 diabetes loci with LADA. Of type 2 diabetes–associated genes, the CC/CT genotypes of rs7961581 (TSPAN8) and the obesity-linked AA/AC genotypes of rs8050136 (FTO) were associated with LADA in general, but mainly in low anti-GAD LADA patients (P = 0.004 and P = 0.004, respectively).
CONCLUSIONS
Genetic heterogeneity in LADA is linked to various degrees of autoimmune activity and may be partly distinct from both type 1 and type 2 diabetes.
Latent autoimmune diabetes in adults (LADA) is a slowly progressive form of autoimmune-associated diabetes (1). It is a common form of diabetes; for example, in the second Nord-Tr⊘ndelag Health Study (HUNT2), 9% of diabetic patients were classified as LADA (2). However, the etiology of LADA remains less well understood than that of other forms of diabetes.
It has been discussed whether LADA is a mild form of type 1 diabetes or a distinct etiological entity (3–5). Previous studies reported that LADA shares genetic features with type 1 diabetes, including an increased frequency of the HLA-DQB1 allele of the DQB1 gene (6,7). However, other results show that age, obesity, and physical inactivity are important risk factors for both LADA and type 2 diabetes (8). Furthermore, the type 2 diabetes–associated gene TCF7L2 has been reported to be associated with LADA (9).
Thus, the etiology of LADA resembles partly type 1 and partly type 2 diabetes. However, the genetic participation of type 1 and type 2 diabetes susceptibility genes in the etiology of LADA needs further elucidation and is complicated by the phenotype of LADA being heterogeneous among and within populations (5). The diagnosis of LADA is usually based on the following: age older than 35 years, the presence of at least one circulating autoantibody against islet cell antigens (usually antibodies to GAD [anti-GAD]), and no apparent need for insulin for at least 6 months after diagnosis. These criteria leave room for heterogeneity. It has, for instance, been reported (10) that the phenotype of individual LADA patients is related to titer of anti-GAD, and in a recent study the need for insulin treatment over several years was more frequent in those with higher titers of anti-GAD (11).
In the present study, we first investigated the association of type 1 and type 2 diabetes candidate loci in LADA patients in general. Second, we tested for variability in genetic background in relation to a marker of autoimmune activity (anti-GAD) as well as a phenotypic risk factor for type 2 diabetes (BMI).
RESEARCH DESIGN AND METHODS
The HUNT2 study population.
The second HUNT study, a population-based study in Nord-Tr⊘ndelag County, was performed between 1995 and 1997. Nord-Tr⊘ndelag is located in the central part of Norway and is fairly demographically representative of Norway, making it suitable for epidemiological studies. All inhabitants ≥20 years (n = 92,936) were invited to participate. The overall response rate was 71.3% (n = 65,258). The survey included a clinical examination, blood sampling, and two general questionnaires that included more than 200 health-related items. In addition, a specific questionnaire was administered to those who stated they had diabetes. Further details are described elsewhere (12).
Identification of diabetic patients.
Diabetic patients were identified by self-reported answer of “yes” to the question “Do you have or have you had diabetes?” (n = 1972). Subjects answering “yes” were invited to a follow-up examination where fasting C-peptide and anti-GAD were measured. In total, 73.6% participated (n = 1,451). At follow-up, participants were also interviewed by the screening nurses to ensure year of diagnosis and details on start of different types of diabetes treatment (12).
C-peptide and anti-GAD measurements.
Serum levels of C-peptide and anti-GAD were analyzed at the Hormone Laboratory of Aker University Hospital, Oslo, Norway. C-peptide was measured by radioimmunoassay (Diagnostic System Laboratories, Webster, Texas) and anti-GAD by immunoprecipitation using [3H] leucine translation labeled GAD65 (Novo Nordisk Pharma AS, Bagsvaerd, Denmark). The anti-GAD assay was based on a validated method (13). Anti-GAD levels were expressed as an index value relative to a standard serum, and a value ≥0.08 was considered positive. At the cutoff level of 0.08, the assay was previously shown (2) by the Diabetes Autoantibody Standardization Program (DASP) to have a sensitivity of 0.64 and specificity of 1.00.
Classification of diabetes.
Patients starting insulin treatment within 6 months of diagnosis were classified as type 1 diabetes if, in addition, they were either anti-GAD positive or anti-GAD negative and had fasting C-peptide levels <150 pmol/l. Patients were classified as LADA if they were anti-GAD positive and had not been treated with insulin within 12 months of diagnosis. Type 2 diabetic case subjects were anti-GAD negative and without insulin treatment within 1 year of diagnosis. By these criteria, we included 120 type 1 diabetic, 126 LADA, and 1,090 type 2 diabetic patients in the present study. One hundred fifteen patients did not meet any of the criteria and were excluded.
Classification of diabetic subjects not attending the follow-up examination.
Not all the identified diabetic case subjects in HUNT2 attended the follow-up examination (n = 519). Nonattendees could therefore not be classified by the criteria given above. Blood and serum samples in nonfasting state were, however, available for 432 of these individuals in the HUNT Biobank. Measuring anti-GAD from the serum samples combined with information on age at diagnosis enabled us to classify these subjects by less stringent criteria (i.e., as type 1 diabetic if anti-GAD positive and age at diagnosis ≤35 years, as LADA if anti-GAD positive and age at diagnosis >35 years and as type 2 diabetic if anti-GAD negative and age of diagnosis >35 years). By these criteria, 16 subjects were classified as type 1 diabetic, 18 as LADA, and 255 as type 2 diabetic patients. One hundred forty-three patients did not meet any of these criteria and were therefore excluded.
Control subject group.
Participants serving as control subjects (n = 1,503) were drawn from the same study. They answered “no” to the question of having diabetes. They were frequency matched by sex and age (in years, by decade) to the diabetic patients.
DNA extraction.
DNA for genotyping was extracted from peripheral blood leukocytes from EDTA whole blood or blood clots using the Gentra Purgene blood kit (QIAGEN Science, Germantown, MD). This was done manually or by automation with an Autopure LS (QIAGEN Science) mainly as described by the manufacturer.
Genotyping.
The selected single nucleotide polymorphisms (SNPs) were based on publicly available results from type 1 (14–20) and type 2 (21–27) diabetes studies. Genotyping was performed by applying TaqMan SNP allelic discrimination using ABI 7900HT and by SNPlex genotyping system (Applied Biosystems, Foster City, CA). Case and control subjects were equally distributed with four or more negative control subjects on each 384-plate. Criteria to pass the assay were 1) call rates >90%, 2) minor allele frequency >1% in the genotyped population, and 3) agreement with Hardy-Weinberg equilibrium in the whole population, and if P value was <0.001 the assay did not pass. Assays that did not pass quality control were excluded from further analysis.
HLA haplotyping.
HLA haplotyping was performed as described by de Bakker et al. (28). They captured nearby single tag SNPs or haplotypes of combination of up to three SNPs as a predictor of known HLA alleles. The recommended tag SNPs or haplotypes for the HLA risk alleles for type 1 diabetes are shown in supplementary Table 1 in the online appendix, which is available at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0923/DC1. We genotyped these tag SNPs using SNPlex genotyping system.
Statistical analysis.
PLINK software (http://pngu.mgh.harvard.edu/∼purcell/plink/index.shtml) was used to assess whether genotypes were in Hardy-Weinberg equilibrium and to test differences in genotype distribution between affected and unaffected subjects by logistic regression under additive, dominant, and recessive models. Odds ratios and 95% CIs were calculated. Adjustment for diabetes-specific risk factors such as age, sex, and BMI was applied when appropriate. Correction for multiple testing was done by max(T) permutation where 1,000 permutations were performed. The nonparametric Mann-Whitney U test was used to compare continuous variables between groups using Statistical Package for the Social Sciences, version 14.0 (SPSS, Chicago). A P value < 0.05 was considered statistically significant. Phasing HLA haplotypes and testing for association within case and control subjects were carried out using PLINK.
Ethics.
All participants gave written consent. The study was approved by the Regional Committee for Ethics in Medical Research and the Norwegian Data Inspectorate.
RESULTS
Clinical characteristics.
Age at onset was similar for LADA and type 2 diabetic patients and lower for type 1 diabetic patients (Table 1). Overweight status, measured by BMI and waist-to-hip ratios, was more marked in LADA and type 2 diabetic than in type 1 diabetic and control subjects. Relationships were similar for lipid parameters and blood pressure.
TABLE 1.
Phenotypic characteristics for male and female subjects selected from the HUNT cohort
| LADA |
Type 1 diabetes |
Type 2 diabetes |
Control subjects |
|||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | |
| n | 68 | 58 | 72 | 48 | 534 | 556 | 740 | 763 |
| Age when attended (years) | 67 ± 12 | 70 ± 11 | 46 ± 16 | 52 ± 17 | 67 ± 11 | 69 ± 11 | 64 ± 15 | 68 ± 14 |
| Age at diagnosis (years) | 58 ± 11 | 60 ± 12 | 26 ± 16 | 34 ± 16 | 60 ± 11 | 61 ± 12 | ||
| BMI (kg/m2) | 27.4 ± 3.8 | 29.4 ± 5.3 | 25.8 ± 3.4 | 26.6 ± 4.6 | 28.5 ± 3.8 | 30.8 ± 5.4 | 26.4 ± 3.4 | 27.1 ± 4.5 |
| Waist-to-hip ratio | 0.94 ± 0.06 | 0.85 ± 0.06 | 0.90 ± 0.06 | 0.80 ± 0.06 | 0.94 ± 0.06 | 0.86 ± 0.7 | 0.91 ± 0.06 | 0.82 ± 0.06 |
| Cholesterol (mmol/l) | 5.4 ± 1.1 | 6.3 ± 1.3 | 5.2 ± 1.0 | 6.0 ± 1.2 | 6.0 ± 1.2 | 6.5 ± 1.3 | 6.0 ± 1.1 | 6.6 ± 1.4 |
| HDL cholesterol (mmol/l) | 1.1 ± 0.4 | 1.4 ± 0.5 | 1.5 ± 0.4 | 1.7 ± 0.5 | 1.1 ± 0.4 | 1.3 ± 0.4 | 1.3 ± 0.4 | 1.5 ± 0.4 |
| Triglycerides (mmol/l) | 2.2 ± 1.4 | 2.3 ± 1.3 | 1.4 ± 0.71 | 1.3 ± 0.73 | 2.6 ± 1.7 | 2.7 ± 1.4 | 1.9 ± 1.1 | 1.9 ± 1.1 |
| Systolic blood pressure (mmHg) | 154 ± 21 | 154 ± 26 | 140 ± 20 | 141 ± 25 | 152 ± 23 | 160 ± 24 | 146 ± 21 | 151 ± 26 |
| Diastolic blood pressure (mmHg) | 85 ± 12 | 80 ± 14 | 79 ± 10 | 78 ± 12 | 86 ± 13 | 86 ± 14 | 84 ± 12 | 83 ± 14 |
| Nonfasting glucose (mmol/l) | 11 ± 5.1 | 10.4 ± 4.6 | 11 ± 6.2 | 10.6 ± 5.5 | 9.8 ± 3.9 | 9.1 ± 4.0 | 5.6 ± 1.3 | 5.7 ± 1.9 |
Data are means ± SD.
Quality testing of SNPs.
Forty-two prioritized SNPs known to be associated with either type 1 or type 2 diabetes were genotyped on either TaqMan or SNPlex in 1,536 diabetic patients (106 additional patients were genotyped [TaqMan] for rs231775, rs689, rs2476601, and rs3118470) from whom genomic DNA was available and in 1,503 nondiabetic control subjects. All SNPs were in Hardy-Weinberg equilibrium (P ≥ 0.001) but two SNPs had genotype call frequency <90% and were therefore excluded from further analysis. The average call frequency for the remaining SNPs was 98.6%.
A full statistical analysis including additive, dominant, and recessive models of all the genotyped type 1 and type 2 diabetes risk loci is shown in supplementary Table 2 for type 1 diabetic compared with control subjects, in supplementary Table 3 for LADA compared with control subjects, and in supplementary Table 4 for type 2 diabetic compared with control subjects.
LADA in general: lack of association of non-HLA type 1 diabetes loci.
We analyzed 12 SNPs within eight candidate risk loci for type 1 diabetes for association with 126 LADA patients. The CC genotype of rs3118470 in the interleukin 2 receptor, alpha gene (IL2Ralfa) was associated with LADA (P = 0.036), but the association became less significant (P = 0.054) after adjustment for age, sex, and BMI (Table 2). None of the other studied type 1 diabetes loci was associated with LADA.
TABLE 2.
Genotypes of known type 1 diabetes–associated loci in LADA and type 1 diabetic subjects compared with nondiabetic control subjects
| Gene name, SNP | Genotype | LADA vs. control subjects |
Type 1 diabetic vs. control subjects |
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P* | P† | P‡ | OR (95% CI) | P* | P† | P‡ | ||
| PTPN22, rs2476601 | AA/AG GG | 1.40 (0.92–2.12) | 0.117 | 0.157 | 1 | 2.82 (1.92–4.13) | 1.17 × 10−7 | 1.46 × 10−5 | 0.001 |
| PTPN22, rs2488457 | CC/CG GG | 1.39 (0.95–2.03) | 0.088 | 0.134 | 1 | 1.70 (1.15–2.50) | 0.008 | 0.034 | 0.851 |
| IL2R, rs3118470 | CC CT/TT | 1.62 (1.03–2.54) | 0.036 | 0.054 | 1 | 1.09 (0.66–1.82) | 0.732 | 0.614 | 1 |
| INS, rs689 | TT AA/AT | 1.10 (0.76–1.59) | 0.626 | 0.598 | 1 | 2.44 (1.60–3.71) | 3.54 × 10−5 | 6.91 × 10−5 | 0.005 |
| INS, rs3842753 | CC AA/AC | 1.18 (0.80–1.74) | 0.396 | 0.368 | 1 | 2.38 (1.54–3.66) | 8.63 × 10−5 | 1.89 × 10−4 | 0.011 |
*P value from logistic regression assuming dominant or recessive model.
†Adjusted P value from logistic regression for age, sex, and BMI.
‡Empirical P value corrected for multiple testing by 1,000 permutations.
LADA in general: association with type 2 diabetes loci.
For type 2 diabetes risk loci, we analyzed 28 SNPs within 25 loci for association with LADA. The CC/CT genotypes of rs7961581 upstream of the tetraspanin 8 gene (TSPAN8) and the obesity-linked AA/AC genotypes of rs8050136 in the fat mass and obesity–associated gene (FTO) were associated with LADA (P = 0.007 and P = 0.005, respectively) as well as type 2 diabetes (P = 0.009 and P = 0.005, respectively) (Table 3). Both loci remained significant after adjusting for age, sex, and BMI.
TABLE 3.
Genotypes of known type 2 diabetes–associated loci in LADA and type 2 diabetic subjects compared with nondiabetic control subjects
| Gene name, SNP | Genotype | LADA vs. control subjects |
Type 2 diabetic vs. control subjects |
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P* | P† | P‡ | OR (95% CI) | P* | P† | P‡ | ||
| TCF7L2, rs7903146 | CT/TT CC | 1.25 (0.85–1.83) | 0.252 | 0.141 | 1 | 1.75 (1.48–2.05) | 1.89 × 10−11 | 4.84 × 10−14 | 0.001 |
| TSPAN8/LGR5, rs7961581 | CC/CT TT | 1.68 (1.15–2.46) | 0.007 | 0.01 | 0.443 | 1.24 (1.06–1.46) | 0.009 | 0.033 | 0.869 |
| FTO, rs8050136 | AA/AC CC | 1.94 (1.22–3.09) | 0.005 | 0.005 | 0.255 | 1.28 (1.08–1.53) | 0.005 | 0.005 | 0.291 |
| FTO, rs1861866 | CT/TT CC | 2.55 (1.45–4.50) | 0.001 | 0.003 | 0.149 | 1.21 (1.00–1.46) | 0.047 | 0.091 | 0.997 |
| FTO, rs9931494 | CG/GG CC | 2.72 (1.63–4.53) | 1.27 × 10−4 | 2.43 × 10−4 | 0.016 | 1.29 (1.09–1.54) | 0.004 | 0.008 | 0.396 |
*P value from logistic regression assuming dominant model.
†Adjusted P value from logistic regression for age, sex, and BMI.
‡Empirical P value corrected for multiple testing by 1,000 permutations.
Verification of associated SNPs.
To verify associations with LADA, we analyzed polymorphisms flanking the SNPs showing an association. We analyzed four additional SNPs in the FTO gene (rs4389136 and rs1861866, 68 kb and 12 kb downstream, respectively, and rs9931494 and rs2388405, 11 kb and 60 kb upstream, respectively) and two SNPs in TSPAN8 (rs7306184, 45 kb downstream and rs7964431, 43 kb upstream) and in IL2R (rs9663421, 46 kb downstream and rs4747880, 51 kb upstream). We selected SNPs with essentially similar minor allele frequencies as the SNPs initially showing an association. Two SNPs (rs7306184 and rs4747880) were excluded from further analyses due to low genotype call rate (<90%).
The GG/GC genotypes of rs1861866 and TT/TC of rs9931491 in the FTO gene were associated with both LADA (P = 0.001 and P = 1.0 × 10−4, respectively) and type 2 diabetes (P = 0.047 and P = 0.004, respectively), and were still associated with LADA after adjustment for age, sex, and BMI (Table 3). The association of rs9931491 with type 2 diabetes remained strong after adjustment for age, sex, and BMI, however, the association of rs1861866 was lost. The SNP rs9931491 in the FTO gene was the only SNP that remained significantly associated with LADA after adjusting for multiple testing with 1,000 permutations (P = 0.016). None of the other SNPs tested was associated with LADA (supplementary Tables 2–4).
Association of genes in relation to BMI.
We dichotomized patients and control subjects on the basis of BMI < (nonobese) or > (obese) 30 kg/m2. The AA/AC genotypes of rs8050136 in the FTO gene were significantly associated with obese LADA patients (P = 0.024) (supplementary Table 5). The two additional SNPs in the FTO gene, the TT/TC genotypes of rs1861866 and the GG/GC genotypes of rs9931494, were associated with both obese (P = 0.048 and P = 0.008, respectively) and nonobese (P = 0.024 and P = 0.017, respectively) LADA patients. However, the association of rs1861866 to obese LADA patients became less significant (P = 0.054) after adjusting for age, sex, and BMI.
The CC/CT genotypes of rs7961581 close to the TSPAN8 gene were associated with nonobese LADA patients (P = 0.003).
Association of anti-GAD titers with genes and clinical characteristics.
We dichotomized LADA on the basis of anti-GAD levels below (low anti-GAD) or above (high anti-GAD) the median of anti-GAD. Regarding the FTO gene, the AA/AC genotypes of rs8050136, TT/TC genotypes of rs1861866, and GG/GC genotypes of rs9931494 were significantly associated with low anti-GAD LADA (P = 0.004, P = 0.004, and P = 0.002, respectively) (supplementary Table 6). In addition, SNP rs9931494 was also associated with high anti-GAD LADA (P = 0.020, supplementary Table 7), but became less significant (P = 0.047) after adjusting for age, sex, and BMI.
The genotypes CC/CT of rs7961581 close to the TSPAN8 gene were associated with low anti-GAD LADA (P = 0.004).
Among clinical characteristics (Table 4), LADA patients with high anti-GAD were younger at diagnosis (P = 0.004) and displayed shorter time to insulin treatment (P = 0.048). Further, fasting glucose was more elevated (P = 0.025) and fasting C-peptide more depressed (P = 0.0007) than in patients with low anti-GAD.
TABLE 4.
Clinical characteristics of LADA patients displayed as low and high titers of anti-GAD
| Clinical characteristics | Patients compared (n) |
Median (25th–75th percentile) |
P* | ||
|---|---|---|---|---|---|
| Low anti-GAD | High anti-GAD | Low anti-GAD | High anti-GAD | ||
| Age at diagnosis (years) | 65 | 61 | 62 (56–71) | 56 (46–66) | 0.004 |
| Years with insulin treatment | 16 | 27 | 6.0 (1.3–9.8) | 6.0 (4.0–11) | 0.605 |
| Time to insulin dependence (years) | 16 | 26 | 6.8 (4.3–12) | 4.0 (2.9–7.0) | 0.048 |
| BMI (kg/m2) | 65 | 58 | 28 (26–31) | 27 (25–31) | 0.182 |
| Waist-to-hip ratio | 64 | 59 | 0.90 (0.85–0.96) | 0.90 (0.85–0.93) | 0.457 |
| Cholesterol (mmol/l) | 65 | 61 | 5.8 (5.2–6.7) | 5.6 (4.8–6.6) | 0.277 |
| HDL cholesterol (mmol/l) | 65 | 61 | 1.2 (0.9–1.5) | 1.1 (0.9–1.5) | 0.870 |
| Triglycerides (mmol/l) | 65 | 61 | 2.27 (1.22–2.98) | 1.77 (1.12–2.66) | 0.158 |
| A1C | 62 | 60 | 7.8 (6.4–9.4) | 8.7 (7.1–10) | 0.064 |
| Systolic blood pressure (mmHg) | 65 | 60 | 154 (143–174) | 148 (132–170) | 0.124 |
| Diastolic blood pressure (mmHg) | 65 | 60 | 82 (72–91) | 81 (73–89) | 0.861 |
| Fasting glucose (mmol/l) | 65 | 61 | 7.5 (6–8.9) | 9.1 (6.9–10.6) | 0.025 |
| Fasting C-peptide (mmol/l) | 65 | 61 | 697 (384–1,015) | 334 (75–764.5) | 0.0007 |
Data are median (25th–75th percentile) unless otherwise indicated.
*P value from Mann-Whitney U test by comparing low and high anti-GAD LADA patients.
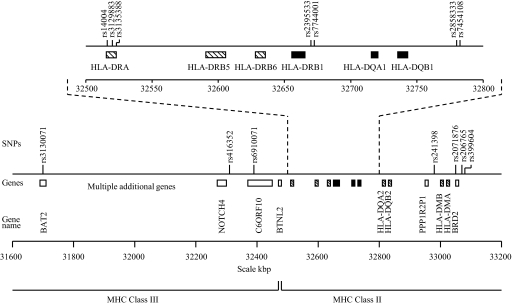
HLA haplotyping.
Of 30 tag SNPs for HLA haplotyping, 18 were successfully genotyped. Of these, 14 could be used in the prediction of HLA haplotypes (supplementary Table 8). The localization of these SNPs in the major histocompatibility complex (MHC) is depicted in Figure 1. The majority of the strongly associated risk haplotypes for type 1 diabetes were also found to be significant risk haplotypes for LADA (supplementary Table 9). The frequency of ACCCG haplotype for DRB1*0402-DQA1*0301-DQB1*0302 was increased in both LADA and type 1 diabetic patients compared with control subjects (0.16 and 0.32 vs. 0.12, P = 0.036 and P = 3.9 × 10−17, respectively) (Table 5). The same was apparent with the GCCG haplotype for DRB1*0401-DQA1*0301-DQB1*0302 (0.16 and 0.33 vs. 0.11, P = 0.028 and P = 7.5 × 10−21), CT haplotype for DRB1*0901-DQA1*0301-DQB1*0303 (0.23 and 0.36 vs. 0.16, P = 0.009 and P = 9.2 × 10−14), AC haplotype for DRB1*1,501-DQA1*0102-DQB1*0602 (0.41 and 0.5 vs. 0.33, P = 0.017 and P = 5.3 × 10−7), and TAT haplotype for DRB1*0701-DQA1*0201-DQB1*0303 (0.12 and 0.25 vs. 0.07, P = 0.015 and P = 2.3 × 10−18). Interestingly, one distinct haplotype GCA for DRB1*0401-DQA1*0301-DQB1*0301 was found to be associated with higher risk only for LADA (0.09 vs. 0.05, P = 0.013).
FIG. 1.
Schematic diagram of the tag SNPs used for HLA haplotyping in the MHC region on chromosome 6 including nearby located genes. The HLA genes that are predicted by the tag SNPs are shown as black boxes; other HLA genes are shown as hatched boxes. Additional genes are shown as white boxes.
TABLE 5.
Frequency of the HLA haplotypes associated with higher risk in LADA and type 1 diabetes
| Risk HLA haplotypes | Type 1 diabetes (n = 119) |
LADA |
||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 121) |
Anti-GAD >0.11 (n = 56) |
Anti-GAD ≤0.11 (n = 65) |
||||||
| Frequency (n) | P* | Frequency (n) | P* | Frequency (n) | P* | Frequency (n) | P* | |
| DRB1*0402-DQA1*0301-DQB1*0302 | ||||||||
| ACCCG | 0.32 (38) | 3.90 × 10−17 | 0.16 (20) | 0.036 | 0.22 (13) | 0.001 | 0.11 (7) | 0.939 |
| AACTG | 0.34 (40) | 0.023 | 0.33 (40) | 0.065 | 0.37 (20) | 0.037 | 0.3 (19) | 0.527 |
| DRB1*0401-DQA1*0301-DQB1*0302 | ||||||||
| GCTA | 0.02 (3) | 0.847 | 0.04 (5) | 0.107 | 0.01 (1) | 0.341 | 0.07 (5) | 0.002 |
| GCCG | 0.33 (40) | 7.48 × 10−21 | 0.16 (19) | 0.028 | 0.21 (12) | 0.001 | 0.11 (7) | 0.926 |
| DRB1*0901-DQA1*0301-DQB1*0303 | ||||||||
| CT | 0.36 (43) | 9.23 × 10−14 | 0.23 (27) | 0.009 | 0.31 (17) | 4.81 × 10−5 | 0.15 (10) | 0.888 |
| DRB1*1,501-DQA1*0102-DQB1*0602 | ||||||||
| AC | 0.50 (60) | 5.31 × 10−7 | 0.41 (50) | 0.017 | 0.41 (23) | 0.105 | 0.42 (27) | 0.069 |
| DRB1*0701-DQA1*0201-DQB1*0303 | ||||||||
| TAT | 0.25 (30) | 2.33 × 10−18 | 0.12 (14) | 0.015 | 0.17 (10) | 0.0002 | 0.07 (5) | 0.962 |
| DRB1*0401-DQA1*0301-DQB1*0301 | ||||||||
| GCA | 0.04 (5) | 0.571 | 0.09 (11) | 0.013 | 0.04 (2) | 0.543 | 0.14 (9) | 7.51 × 10−5 |
| GCG | 0.36 (43) | 1.97 × 10−10 | 0.20 (25) | 0.483 | 0.23 (13) | 0.305 | 0.18 (12) | 0.986 |
| DRB1*0403-DQA1*0301-DQB1*0302 | ||||||||
| CCGTG | 0.13 (15) | 7.57 × 10−12 | 0.05 (6) | 0.179 | 0.08 (4) | 0.016 | 0.03 (2) | 0.692 |
*P value corrected for age, sex, and BMI.
In general, the most strongly associated protective haplotypes for type 1 diabetes were also found to be protective in LADA (supplementary Table 10). The frequency of ATTG haplotype for DRB1*0401-DQA1*0301-DQB1*0302 was significantly lower in both LADA and type 1 diabetic patients compared with control subjects (0.18 and 0.06 vs. 0.25, P = 0.014 and P = 2.9 × 10−10, respectively) (Table 6). The same was observed for the TC haplotype for DRB1*0902-DQA1*0301-DQB1*0303 (0.15 and 0.06 vs. 0.24, P = 0.003 and P = 1.5 × 10−9), CT haplotype for DRB1*1,501-DQA1*0102-DQB1*0602 (0.09 and 0.03 vs. 0.15, P = 0.025 and P = 1.7 × 10−6), and ATG haplotype for DRB1*0401-DQA1*0301-DQB1*0301 (0.12 and 0.04 vs. 0.19, P = 0.005 and P = 8.3 × 10−9).
TABLE 6.
Frequency of the HLA haplotypes associated with protective effect in LADA and type 1 diabetes
| Protective HLA haplotypes | Type 1 diabetes (n = 119) |
LADA |
||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 121) |
Anti-GAD >0.11 (n = 56) |
Anti-GAD ≤0.11 (n = 65) |
||||||
| Frequency (n) | P* | Frequency (n) | P* | Frequency (n) | P* | Frequency (n) | P* | |
| DRB1*0402-DQA1*0301-DQB1*0302, TCTTG | 0.03 (3) | 0.0003 | 0.07 (9) | 0.092 | 0.04 (2) | 0.031 | 0.1 (7) | 0.731 |
| DRB1*0401-DQA1*0301-DQB1*0302, ATTG | 0.06 (7) | 2.94 × 10−10 | 0.18 (21) | 0.014 | 0.14 (8) | 0.012 | 0.21 (14) | 0.288 |
| DRB1*0901-DQA1*0301-DQB1*0303, TC | 0.06 (7) | 1.51 × 10−9 | 0.15 (18) | 0.003 | 0.11 (6) | 0.003 | 0.19 (12) | 0.203 |
| DRB1*1,501-DQA1*0102-DQB1*0602, CT | 0.03 (4) | 1.73 × 10−6 | 0.09 (11) | 0.025 | 0.06 (3) | 0.014 | 0.12 (8) | 0.418 |
| DRB1*0701-DQA1*0201-DQB1*0303, TGC | 0.2 (24) | 7.67 × 10−6 | 0.29 (35) | 0.123 | 0.24 (13) | 0.027 | 0.34 (22) | 0.922 |
| DRB1*0401-DQA1*0301-DQB1*0301, ATG | 0.04 (4) | 8.28 × 10−9 | 0.12 (14) | 0.005 | 0.1 (6) | 0.017 | 0.13 (9) | 0.093 |
| DRB1*0403-DQA1*0301-DQB1*0302, TTGTA | 0.02 (2) | 1.46 × 10−5 | 0.09 (11) | 0.349 | 0.07 (4) | 0.167 | 0.11 (7) | 0.993 |
*P value corrected for age, sex, and BMI.
When dichotomizing LADA for high and low anti-GAD, both the risk and protective HLA haplotypes described above were now found to be associated with high anti-GAD LADA patients (Tables 5 and 6), except the GCA haplotype for DRB1*0401-DQA1*0301-DQB1*0301, which was associated with higher risk in low anti-GAD LADA patients (P = 0.0003). Surprisingly, one more distinct haplotype was found. The GCTA haplotype for DRB1*0401-DQA1*0301-DQB1*0302 was associated with higher risk only in low anti-GAD LADA (P = 0.006) and not with type 1 and type 2 diabetes.
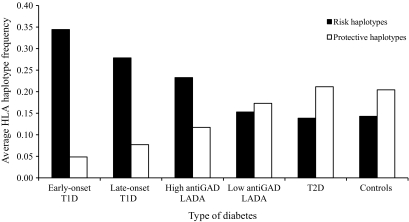
Age at diagnosis of type 1 diabetes correlates with HLA haplotype frequencies (29). Dichotomizing the type 1 diabetic patients on the basis of age of diagnosis at older than or younger than 35 years enabled us to compare type 1 and LADA patients on a more equal age-at-onset basis. We selected the average frequency from the seven strongest associated risk HLA haplotypes and the average frequency from the seven most protective HLA haplotypes for type 1 diabetes to illustrate the differences in HLA frequencies in different types of diabetes (Fig. 2). Risk haplotype frequencies tended to decrease and protective haplotype frequencies to increase when comparing patients and control subjects in the following order: early-onset type 1 diabetic to late-onset type 1 diabetic through high anti-GAD LADA and low anti-GAD LADA to type 2 diabetic and control subjects. This suggests a continuous spectrum from strong to weak to nonexistent influence of autoimmunity.
FIG. 2.
Average HLA haplotype frequencies in different types of diabetes, calculated from the seven strongest associated risk HLA haplotypes and the seven strongest protective HLA haplotypes with type 1 diabetes. T1D, type 1 diabetes; T2D, type 2 diabetes.
Including less stringently classified patients.
The clinical characteristics for the 18 patients who were classified as LADA by less stringent criteria were mainly the same as patients characterized from the complete dataset (supplementary Table 11). After including them in the analysis, the type 1 diabetes–associated CC/CG genotypes of rs2488457 in the protein tyrosine phosphatase nonreceptor type 22 gene (PTPN22), CC genotype of rs2296336 in the inositol 1,4,5-triphosphate receptor type 3 gene (ITPR3), and the CC genotype of rs3118470 in the IL2R gene were associated with LADA (P = 0.039, P = 0.008, and P = 0.008, respectively) (data not shown). The SNPs rs2296336 and rs3118470 remained significant after adjusting for age, sex, and BMI (P = 0.015 and P = 0.019, respectively), however the rs2488457 showed only a trend (P = 0.073) after adjustment. The associations to the type 2 diabetes loci TSPAN8 and FTO remained significant, however, the associations were slightly weakened. There were no differences in the association of the FTO and TSPAN8 genes to the dichotomized LADA patients. However, the type 1 diabetes–associated loci PTPN22 (rs2476601), ITPR3 (rs2296336), and IL2R (rs3118470) were now associated with nonobese LADA patients (P = 0.039, P = 0.015, and P = 0.029, respectively) and high anti-GAD levels (P = 0.034, P = 0.037, and P = 0.011, respectively). The associated HLA haplotypes with LADA became even stronger.
DISCUSSION
Our study indicates that in LADA patients 1) admixture of both type 1 and type 2 diabetes–associated genetic variants are present, 2) heterogeneity is related to autoimmune activity as assessed by anti-GAD and insulin resistance as assessed by BMI, and 3) there is suggestive evidence for genetic associations that are not found in either type 1 or type 2 diabetes.
Admixture of type 1 diabetes genes in LADA was apparent for HLA haplotypes that confer risk for or protection against type 1 diabetes. These findings are in agreement with previous studies (9,30). In general, the frequency of these haplotypes as well as significance levels were more pronounced for type 1 diabetes than for LADA, although differences were attenuated when comparing late age-at-onset type 1 diabetes and LADA (supplementary Tables 9 and 10). On the other hand, type 1 diabetes genes outside the HLA region were not in apparent association with LADA. We found, as reported (31), strong associations between type 1 diabetes and SNPs rs689 and rs3842753 in the insulin gene, whereas such associations were lacking in LADA. Similar difference between type 1 diabetes and LADA were also noted for the genes PTPN22 and CTLA4 (supplementary Tables 2 and 3).
In regards to admixture of genes associated with type 2 diabetes, there was correspondence with LADA for the FTO and TSPAN8/LGR5 genes. The association with FTO and type 2 diabetes in the HUNT2 population was previously reported (32), whereas our finding in LADA patients is novel. Notably, both the associations in type 2 diabetes and in LADA patients remained after adjustment for BMI. Such adjustment did in most (21,33) but not all (32,34) other studies abolish the association with type 2 diabetes. The high expression of the FTO gene in hypothalamus and regulation by food intake suggests a role in controlling energy homeostasis (35,36). However, underlying mechanisms of influence in obesity and diabetes are not fully elucidated. TSPAN8 is a member of the transmembrane 4 superfamily. Its role in the development of diabetes is still unknown.
The association of FTO with LADA was found mainly in those with low titers of anti-GAD and in those with high BMI. An inverse relationship with autoimmune activity is consistent with the absence of these SNP associations in type 1 diabetes. Also, most of the HLA associations in LADA patients were attenuated at lower versus higher titers of anti-GAD. Relationships with BMI were generally the inverse of those with anti-GAD. Collectively, our data suggest a graded influence of the genetic predisposition from a more type 1 diabetes–like one in those who exhibit high autoimmune activity and lower BMI, toward a more type 2 diabetes–like one in those that exhibit lower titers of anti-GAD and higher BMI.
We did not confirm an association between LADA and the TCF7L2 gene (supplementary Table 3) that was reported by Cervin et al. (9). The definition of LADA in the study of Cervin et al. differs from ours, and there are differences in the anthropometric data. However, we have no clear-cut explanation for the discrepancy. One may argue that our study was underpowered to detect an impact of TCF7L2 in LADA. Still the high level of significance for association in type 2 diabetes (P < 1.0 × 10−10, supplementary Table 4) is in obvious contrast to the lack of significance in LADA. One could envisage that heterogeneity of LADA with regard to autoimmune activity assessed by anti-GAD could obscure an association with TCF7L2 in a subgroup with lower autoimmune activity. However, we could not detect such an influence (supplementary Table 6).
Our results provide some suggestive evidence for genetic risk factors for LADA that are related to neither type 1 nor type 2 diabetes. The IL2R gene showed a trend toward association with LADA but not with type 1 or type 2 diabetes. Also, two distinct HLA haplotypes were associated only with LADA. These observations may serve as pointers for studies that rigorously test the notion of partly unique genetic predisposition for LADA.
A previous HUNT study (2) and a study in the U.K. (37), demonstrated inverse relationships between anti-GAD titer and the odds ratio for effect of family history of diabetes in LADA patients. This would suggest a strong effect of nonautoimmune genes (most of which have yet to been identified) in LADA patients.
Our study has limitations and strengths. An obvious limitation is the relatively low number of patients in the groups classified as type 1 diabetic and LADA. As a consequence, the inability to obtain formal levels of significance for some gene associations may reside in lack of power. Another potential weakness is the lack of complete data on all professed diabetic patients. After retrospectively analyzing anti-GAD in most of these individuals, we could classify them into different forms of diabetes by wider criteria than for those patients who were the main focus of this study. By including these patients in some of our analyses, we could estimate a possible bias occurring from their exclusion. The “new” LADA patients were in genetic terms perhaps more type 1 diabetes–like than the average LADA patients in whom we had access to complete data. However, including the new LADA patients in our analysis did not materially change the conclusions that were based on complete sets of data.
A technical limitation may pertain to anti-GAD measurements, which were not compared with an alternative method. However, our assay has been used and validated previously (13). Furthermore, the outcome of a DASP investigation indicates very high specificity of the assay coupled with a lower sensitivity. The main concern here could then be that we have missed some diabetic subjects who, by other methods, could have been anti-GAD positive.
Strengths of our study pertain to the HUNT studies being all-population inclusive with a high attendance. Notably our population-based group of adult type 1 diabetes is rather unique because most other studies that include type 1 diabetic subjects are based on hospital records. Our data also indicate that the type 1 diabetic subjects are representative outside a regional perspective. Thus, data are in good agreement with published ones on type 1 diabetes with regard to specific risk and protective genes in white subjects and a diminished influence of diabetes-associated HLA haplotypes with increasing age of diagnosis (29). As to our type 2 diabetic and control groups, they have previously been part of a replication in a genome-wide association study (27) and other genetic studies (32,38,39). The results of the HUNT part of these previous studies showed in general good concordance with other patient materials. The evidence that our study is representative of both type 1 and type 2 diabetes should increase the validity of comparisons with LADA.
In summary, we find genetic resemblance to both type 1 and type 2 diabetes genes in LADA. Heterogeneity in susceptibility genes in LADA is related to various degrees of autoimmune activity and obesity. This highlights the concept that it is the combined burden of risk factors that brings on diabetes.
Supplementary Material
Acknowledgments
The HUNT Study is a collaboration among The HUNT Research Centre, Faculty of Medicine (DMF), The Norwegian University of Science and Technology (NTNU), The National Institute of Public Health, and The Nord-Tr⊘ndelag County Council. The current study was supported by the Liaison Committee between the Central Norway Regional Health Authority and NTNU and the Liaison Committee between St. Olav's Hospital Trust and DMF, NTNU.
No potential conflicts of interest relevant to this article were reported.
Parts of this study were presented as a poster at the 45th annual meeting of the European Association for the Study of Diabetes, Vienna, Austria, 29 September–2 October 2009.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Fourlanos S, Dotta F, Greenbaum CJ, Palmer JP, Rolandsson O, Colman PG, Harrison LC: Latent autoimmune diabetes in adults (LADA) should be less latent. Diabetologia 2005;48:2206–2212 [DOI] [PubMed] [Google Scholar]
- 2.Carlsson S, Midthjell K, Grill V: Influence of family history of diabetes on incidence and prevalence of latent autoimmune diabetes of the adult: results from the Nord-Trøndelag Health Study. Diabetes Care 2007;30:3040–3045 [DOI] [PubMed] [Google Scholar]
- 3.Palmer JP, Hampe CS, Chiu H, Goel A, Brooks-Worrell BM: Is latent autoimmune diabetes in adults distinct from type 1 diabetes or just type 1 diabetes at an older age? Diabetes 2005;54(suppl 2):S62–S67 [DOI] [PubMed] [Google Scholar]
- 4.Groop L, Tuomi T, Rowley M, Zimmet P, Mackay IR: Latent autoimmune diabetes in adults (LADA)–more than a name. Diabetologia 2006;49:1996–1998 [DOI] [PubMed] [Google Scholar]
- 5.Gale EA: Latent autoimmune diabetes in adults: a guide for the perplexed. Diabetologia 2005;48:2195–2199 [DOI] [PubMed] [Google Scholar]
- 6.Tuomi T, Carlsson A, Li H, Isomaa B, Miettinen A, Nilsson A, Nissén M, Ehrnström BO, Forsén B, Snickars B, Lahti K, Forsblom C, Saloranta C, Taskinen MR, Groop LC: Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes 1999;48:150–157 [DOI] [PubMed] [Google Scholar]
- 7.Haller K, Kisand K, Pisarev H, Salur L, Laisk T, Nemvalts V, Uibo R: Insulin gene VNTR, CTLA-4 +49A/G and HLA-DQB1 alleles distinguish latent autoimmune diabetes in adults from type 1 diabetes and from type 2 diabetes group. Tissue Antigens 2007;69:121–127 [DOI] [PubMed] [Google Scholar]
- 8.Carlsson S, Midthjell K, Tesfamarian MY, Grill V: Age, overweight and physical inactivity increase the risk of latent autoimmune diabetes in adults: results from the Nord-Trøndelag health study. Diabetologia 2007;50:55–58 [DOI] [PubMed] [Google Scholar]
- 9.Cervin C, Lyssenko V, Bakhtadze E, Lindholm E, Nilsson P, Tuomi T, Cilio CM, Groop L: Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes 2008;57:1433–1437 [DOI] [PubMed] [Google Scholar]
- 10.Lohmann T, Kellner K, Verlohren HJ, Krug J, Steindorf J, Scherbaum WA, Seissler J: Titre and combination of ICA and autoantibodies to glutamic acid decarboxylase discriminate two clinically distinct types of latent autoimmune diabetes in adults (LADA). Diabetologia 2001;44:1005–1010 [DOI] [PubMed] [Google Scholar]
- 11.Radtke MA, Midthjell K, Nilsen TI, Grill V: Heterogeneity of patients with latent autoimmune diabetes in adults: linkage to autoimmunity is apparent only in those with perceived need for insulin treatment: results from the Nord-Trøndelag Health (HUNT) study. Diabetes Care 2009;32:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmen J, Midthjell K, Krüger Ø, Langhammer A, Holmen TL, Bratberg GH, Vatten L, Lund-Larsen PG: The Nord-Trøndelag Health Study 1995–97 (HUNT2): objectives, contents, methods and participation. Norsk Epidemiologi 2003;13:19–32 [Google Scholar]
- 13.Petersen JS, Hejnaes KR, Moody A, Karlsen AE, Marshall MO, Høier-Madsen M, Boel E, Michelsen BK, Dyrberg T: Detection of GAD65 antibodies in diabetes and other autoimmune diseases using a simple radioligand assay. Diabetes 1994;43:459–467 [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki E, Awata T, Ikegami H, Kobayashi T, Maruyama T, Nakanishi K, Shimada A, Uga M, Uga M, Kurihara S, Kawabata Y, Tanaka S, Kanazawa Y, Lee I, Eguchi K: Systematic search for single nucleotide polymorphisms in a lymphoid tyrosine phosphatase gene (PTPN22): association between a promoter polymorphism and type 1 diabetes in Asian populations. Am J Med Genet A 2006;140:586–593 [DOI] [PubMed] [Google Scholar]
- 15.Qu HQ, Montpetit A, Ge B, Hudson TJ, Polychronakos C: Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes 2007;56:1174–1176 [DOI] [PubMed] [Google Scholar]
- 16.Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, Sheridan CM, Glusman G, Hood L, Lernmark A, Janer MSwedish Childhood Diabetes Study Group, Diabetes Incidence in Sweden Study Group Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am J Hum Genet 2006;79:614–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Rønningen KS, Guja C, Ionescu-Tîrgovişte C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC: Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511 [DOI] [PubMed] [Google Scholar]
- 18.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C: A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 2007;448:591–594 [DOI] [PubMed] [Google Scholar]
- 19.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, Savage DA, Walker NM, Clayton DG, Todd JA: A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 2006;38:617–619 [DOI] [PubMed] [Google Scholar]
- 20.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tîrgovişte C, Genetics of Type 1 Diabetes in Finland. Simmonds MJ, Heward JM, Gough SC, Wellcome Trust Case Control Consortium. Dunger DB, Wicker LS, Clayton DG: Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, Wellcome Trust Case Control Consortium (WTCCC) McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 24.Damcott CM, Ott SH, Pollin TI, Reinhart LJ, Wang J, O'connell JR, Mitchell BD, Shuldiner AR: Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the Old Order Amish. Diabetes 2005;54:2245–2250 [DOI] [PubMed] [Google Scholar]
- 25.Meyre D, Bouatia-Naji N, Vatin V, Veslot J, Samson C, Tichet J, Marre M, Balkau B, Froguel P: ENPP1 K121Q polymorphism and obesity, hyperglycaemia and type 2 diabetes in the prospective DESIR Study. Diabetologia 2007;50:2090–2096 [DOI] [PubMed] [Google Scholar]
- 26.Aulchenko YS, Pullen J, Kloosterman WP, Yazdanpanah M, Hofman A, Vaessen N, Snijders PJ, Zubakov D, Mackay I, Olavesen M, Sidhu B, Smith VE, Carey A, Berezikov E, Uitterlinden AG, Plasterk RH, Oostra BA, van Duijn CM: LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes 2007;56:3020–3026 [DOI] [PubMed] [Google Scholar]
- 27.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Boström KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jørgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjögren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Wellcome Trust Case Control Consortium. Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D: Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD: A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006;38:1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabbah E, Savola K, Ebeling T, Kulmala P, Vähäsalo P, Ilonen J, Salmela PI, Knip M: Genetic, autoimmune, and clinical characteristics of childhood- and adult-onset type 1 diabetes. Diabetes Care 2000;23:1326–1332 [DOI] [PubMed] [Google Scholar]
- 30.Desai M, Zeggini E, Horton VA, Owen KR, Hattersley AT, Levy JC, Walker M, Gillespie KM, Bingley PJ, Hitman GA, Holman RR, McCarthy MI, Clark A: An association analysis of the HLA gene region in latent autoimmune diabetes in adults. Diabetologia 2007;50:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavesen MG, McCormack R, Guja C, Ionescu-Tîrgovişte C, Undlien DE, Rønningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Bennett ST, Clayton DG, Cordell HJ, Todd JA: Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes 2004;53:1884–1889 [DOI] [PubMed] [Google Scholar]
- 32.Hertel JK, Johansson S, Raeder H, Midthjell K, Lyssenko V, Groop L, Molven A, Njølstad PR: Genetic analysis of recently identified type 2 diabetes loci in 1,638 unselected patients with type 2 diabetes and 1,858 control participants from a Norwegian population-based cohort (the HUNT study). Diabetologia 2008;51:971–977 [DOI] [PubMed] [Google Scholar]
- 33.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI: A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, Kamboh MI: Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet 2008;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fredriksson R, Hägglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, Levine AS, Lindblom J, Schiöth HB: The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology 2008;149:2062–2071 [DOI] [PubMed] [Google Scholar]
- 36.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O'Rahilly S, Schofield CJ: The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007;318:1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castleden HA, Shields B, Bingley PJ, Williams AJ, Sampson M, Walker M, Gibson JM, McCarthy MI, Hitman GA, Levy JC, Hattersley AT, Vaidya B, Pearson ER: GAD antibodies in probands and their relatives in a cohort clinically selected for Type 2 diabetes. Diabet Med 2006;23:834–838 [DOI] [PubMed] [Google Scholar]
- 38.Johansson S, Raeder H, Eide SA, Midthjell K, Hveem K, Søvik O, Molven A, Njølstad PR: Studies in 3,523 Norwegians and meta-analysis in 11,571 subjects indicate that variants in the hepatocyte nuclear factor 4 alpha (HNF4A) P2 region are associated with type 2 diabetes in Scandinavians. Diabetes 2007;56:3112–3117 [DOI] [PubMed] [Google Scholar]
- 39.Thorsby PM, Midthjell K, Gjerlaugsen N, Holmen J, Hanssen KF, Birkeland KI, Berg JP: Comparison of genetic risk in three candidate genes (TCF7L2, PPARG, KCNJ11) with traditional risk factors for type 2 diabetes in a population-based study–the HUNT study. Scand J Clin Lab Invest 2009;69:282–287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.