Abstract
Type IV secretion systems are secretion nanomachines spanning the two membranes of Gram-negative bacteria. Three proteins, VirB7, VirB9, and VirB10 assemble into a 1.05 MDa core spanning the inner and outer membranes. This core consists of 14 copies of each of the proteins and forms two layers, the I and O layers, inserting in the inner and outer membrane, respectively. Here we present the crystal structure of a ~0.6 MDa outer membrane complex containing the entire O-layer. This structure is the largest determined for an outer membrane channel and is also unprecedented in being composed of three proteins. Unexpectedly, this structure identifies VirB10 as the outer membrane channel with a unique hydrophobic double helical trans-membrane region. This structure establishes VirB10 as the only known protein crossing both membranes of Gram-negative bacteria. Comparison of the cryo-EM and crystallographic structures point to conformational changes regulating channel opening and closing.
Type IV secretion (T4S) systems are utilized by Gram-negative bacteria in a variety of processes, ranging from the delivery of virulence factors into eukaryotic cells to conjugative transfer of genetic material and the uptake or release of DNA1-3. In Helicobacter pylori, Brucella suis and Legionella pneumophila, T4S systems mediate the injection of virulence proteins into mammalian host cells to cause gastric ulcers, brucellosis, or Legionnaire disease, respectively4,5. In A. tumefaciens, the VirB/D T4S system delivers oncogenic DNA and proteins into plant cells2,6 and Bordetella pertussis uses a T4S system to secrete the pertussis toxin into the extracellular milieu7. Conjugation promotes bacterial genome plasticity and the adaptive response of bacteria to changes in the environment, contributing to the spread of antibiotic resistance genes among pathogenic bacteria8.
Although variations exist, many of the T4S systems found in Gram-negative bacteria are similar to the A. tumefaciens VirB/D T4S system, which comprises 12 proteins named VirB1 to VirB11 and VirD42. In general (but not always), T4S systems include an extracellular pilus composed of a major (VirB2) and a minor (VirB5) subunit9. Three ATPases located at the inner membrane, VirB4, VirB11, and VirD4, power substrate secretion and possibly assist in the assembly of the system2,10. The inner membrane channel is thought to be composed of the polytopic membrane protein VirB6 and the bitopic membrane proteins VirB8 and VirB102. At the outer membrane, the composition of the pore that allows the substrate to reach the extracellular milieu is unknown. VirB9 in complex with the short lipoprotein VirB7 could be part of this structure. A region of the C-terminal domain of VirB9 was shown to be surface-exposed11. However, no trans-membrane region could be found or predicted in both proteins.
Recently, the cryo-EM structure of the core complex of the T4S system encoded by the conjugative plasmid pKM101 showed that T4S systems consist of a 1.05 MDa core spanning the inner and outer membranes of Gram-negative bacteria12. This core, extracted pre-assembled from the membranes using detergents, is composed of two layers, the O- and I- layers, and is formed by 14 copies of three proteins, the homologs of the VirB7, VirB9, and VirB10 proteins termed TraN, TraO, and TraF, respectively. Here we present the crystal structure of the T4S system outer membrane complex, containing the entire O-layer.
General architecture of the complex
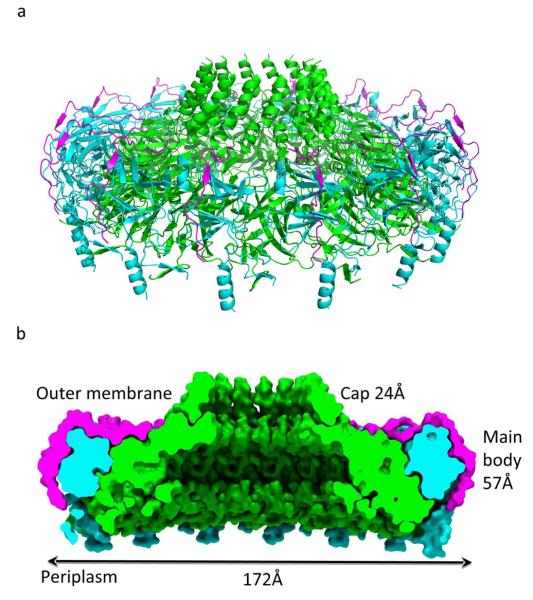
The T4S system outer membrane complex was obtained from chymotryptic cleavage of the T4S system core, crystallized and its structure solved as described in methods section (see also Supplementary Table 1 and Supplementary Fig. 1). This 590 kDa complex contains 14 copies each of the C-terminal domain of TraF (TraFCT; residues 160 to 386), the C-terminal domain of TraO (TraOCT; residues 160 to 294) and the full-length TraN (Supplementary Fig. 1a). The hetero-tetradecameric structure of the T4S system outer membrane complex has an elegant architecture with 14-fold symmetry (Fig. 1a). In the cryo-EM work, the O-layer was described as containing two parts: the main body and the cap. In the crystal structure, the cap is made of a hydrophobic ring of 2-helix bundles defining a 32Å channel (Fig. 1, a and b). These helices are made of residues 307 to 355 of TraFCT. The main body is made of the rest of TraFCT, TraOCT, and TraN. It has a 172Å diameter (Fig. 1b). When the complex is viewed from the top i.e. from the extracellular milieu (Supplementary Fig. 2a), TraFCT forms an inner ring surrounded by the TraOCT-TraN complex. The TraN protein forms spokes radially crossing the entire assembly (Supplementary Figs. 2a and 2b). When the complex is viewed from the bottom i.e. from the periplasm (Supplementary Fig. 2c), the structure is primarily made of TraFCT. A cut-away view of the structure (Fig. 1b) shows that TraF forms the entirety of the inner wall of the structure, establishing VirB10 homologs as T4S system outer membrane channel proteins, an unexpected result as VirB10 homologs have never been suspected to form the outer membrane channel of T4S systems. VirB10 homologs are also found in the inner membrane fraction due to the presence of a trans-membrane helix at the N-terminus of the protein13. Thus, VirB10 is the only protein known to insert in both inner and outer membranes of Gram-negative bacteria.
Figure 1.
The T4S system outer membrane complex. Ribbon diagram (a) and space-filling cut-away (b) of the tetradecameric complex. TraFCT, TraOCT and TraN subunits are colour-coded green, cyan, and magenta, respectively. In b, dimensions and labelling of the various parts of the complex are provided.
Structure of the heterotrimer unit
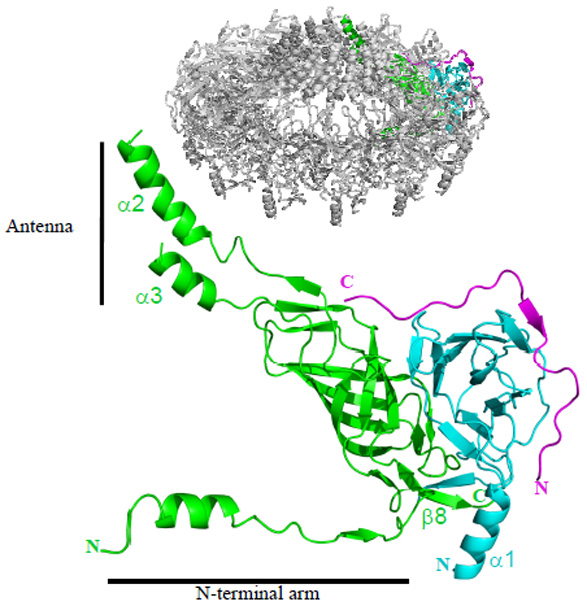
A unique feature of the T4S system outer membrane complex is that it is made of three proteins (instead of one for all outer membrane structures previously determined), all three indispensable for complex assembly and channel formation12. The heterotrimer formed by TraFCT, TraOCT and TraN is shown in Fig. 2 (see also Supplementary Fig. 2d). The interfaces between TraFCT and TraOCT and between TraOCT and TraN bury 2861 Å2 and 2024 Å2 of surface area, respectively. TraN does not interact with TraFCT in the heterotrimer. The core structure of the TraOCT-TraN complex bound to TraFCT is similar to the previously determined NMR structure of the TraOCT-TraN complex alone (Fig. 2; details in supplementary text and Supplementary Figs. 3a and 3b)11. However, there are also important differences. Firstly, in TraOCT bound to TraFCT, residues 162 to 173 at the N-terminus forms the α1 helix (Fig. 2). In the tetradecamer, 14 α1 helices form 14 pillars onto which the entire complex sits on the I-layer (Fig. 1a). Secondly, at the C-terminus, residues 286 to 289 have become ordered in TraOCT bound to TraFCT to form the β10 strand. β10 of TraOCT is part of the TraFCT-TraOCT interface as it forms a 2-stranded β-sheet with β8 of TraFCT (see Fig. 2 where, for clarity, only β8 of TraFCT is indicated). In TraN, sequences at the N- (residues 15 to 25) and C- (residues 37 and 42) termini have become ordered in the heterotrimer. Remarkably, TraN almost entirely wraps around TraOCT, forming an interface with TraOCT that is much more extensive than the one observed in the NMR structure (see details in supplementary text and Supplementary Fig. 3c).
Figure 2.
Ribbon diagram of the heterotrimer unit. Structures mentioned in the main text are indicated. The insert locates the shown heterotrimer within the tetradecameric structure. A stereo version of this figure with full secondary structure labelling is provided in Supplementary Fig. 2d.
The structure of the apo form of the C-terminal domain of ComB10 (ComB10CT), a TraF/VirB10 homolog, has been solved14. It consists of an atypical β-barrel flanked by an α-helix and onto which an “antenna” like structure is mounted (Supplementary Fig. 3d). TraFCT in the heterotrimer differs from ComB10CT in three major ways: i- the flanking helix of ComB10CT is missing in TraFCT; ii- the antenna structure of TraFCT is more extended; and iii- the N-terminus of TraFCT forms an extended N-terminal arm (or lever arm; residues 171 to 199) that projects out to interact with three consecutive neighbouring heterotrimers in the 14mer (Figs. 2 and 3; Supplementary Figs. 3e and 4).
Figure 3.
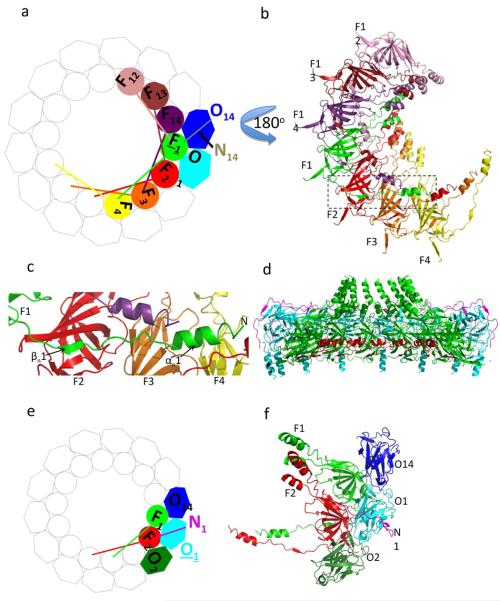
Inter-heterotrimeric interactions. a. Schematic diagram of the tetradecamer with emphasis on interactions with the TraFCT subunit in green (F1). TraFCT and TraOCT subunits are shown in circles and hexagons, respectively. Not shown for clarity: TraN subunits (except N14 that makes interactions with F1) and the lever arms of subunits not interacting with F1. b. Ribbon diagram of the F1-interacting TraFCT subunits (i.e F4, F3, F2, F14, F13, and F12) viewed from the periplasm i.e. turned 180° compared to the view in panel a. The rectangle locates the lever arm of subunit F1. c. Interactions of subunits F2, F3, and F4 with the lever arm of subunit F1. The secondary structures of the lever arm of F1 are labelled. The F2, F3, and F4 subunits are in ribbon representation and labelled accordingly. d. Cut-away side view of the outer membrane complex with the proteins in ribbon diagram representation color-coded as in Fig. 1 but for the N-terminal arms of the TraFCT subunits in red. e. Schematic diagram of the tetradecamer with emphasis on interactions with the TraOCT subunit in cyan (O1). Not shown for clarity: TraN subunits (except N1 that makes interactions with O1) and the lever arms of subunits not interacting with O1. f. Ribbon diagram of the subunits shown in colour at left. This view is from the extracellular milieu.
Tetradecameric assembly
The interactions between heterotrimers in the tetradecameric outer membrane complex are extensive (details are provided in Supplementary Figs. 4, 5, 6 and 7, and supplementary text). To help orientate the reader, a consistent colour scheme summarized in Figs. 3a and 3e is provided, in which the TraF/VirB10, TraO/VirB9 and TraN/VirB7 subunits are numbered clockwise F1 to F14, O1 to O14 and N1 to N14, respectively.
Of the 13000 Å2 of total surface area of TraFCT, 11 % is involved in intra-heterotrimeric interactions and 51.5 % is involved in interactions between heterotrimers (Supplementary Figs. 8a and 8b). TraFCT of one heterotrimer (for example F1 in green in Figs. 3a and 3b) interacts not only with two adjacent TraFCT subunits (F2 and F14), but also, because of the long N-terminal lever arm, with four neighbouring TraFCT subunits located further afield (F3, F4 on one side and F13 and F12 on the other; Fig. 3b). In addition TraFCT interacts with the TraOCT and TraN subunits of an adjacent heterotrimer (O14 and N14 in Fig. 3a)
The interface between TraFCT subunits includes 2320 Å2 of surface area and is described in details in supplementary text and in Supplementary Fig. 6. Strand additions and numerous loop-loop interactions constitute important parts of the interface. Yet, its most notable feature is the N-terminal lever arm of the TraFCT subunit, which makes its own extensive interaction network. The arm of subunit FX (1≤X≤14) interacts with the subunit FX+1, FX+2, and FX+3 (for example, see Figs. 3b and 3c where the F1 lever arm interacts with subunit F2, F3 and F4). FX-FX+1, FX-FX+2, and FX-FX+3 contacts involve residues in the βn1 strand, the βn1-αn1 linker, and αn1 helix of the FX subunit lever arm, respectively (Fig. 3c and Supplementary Figs. 6c and 6d). Overall, the N-terminal lever arms form a continuous inner shelf at the base of the outer membrane complex (Fig. 3d).
In the tetradecameric structure of the outer membrane complex, TraOCT interacts with 5 proteins: two adjacent TraOCT subunits, 1 TraFCT subunit of an adjacent heterotrimer, 1 TraFCT and 1 TraN within its own heterotrimer (Figs. 3e and 3f). Overall, 49.25% of the total surface area of TraOCT is involved in protein-protein interaction, 43% of which is with the adjacent heterotrimer proteins (Supplementary Figs. 8c and 8d). The interfaces between adjacent TraOCT subunits and between TraOCT of one heterotrimer and TraFCT of an adjacent heterotrimer (described in supplementary text and Supplementary Fig. 7) mostly consist of loop residues (Fig. 3f).
The outer membrane pore
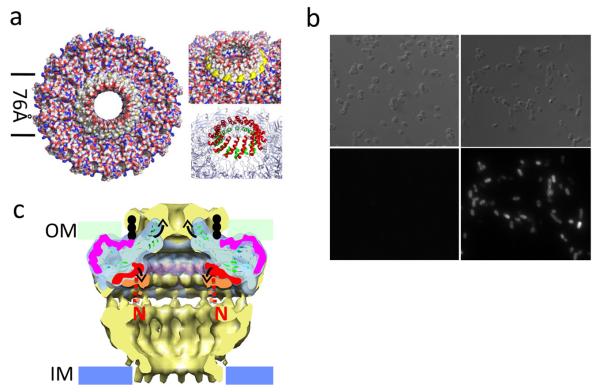
Viewed from the top, the structure contains a central hydrophobic ring of 76 Å in diameter (Fig. 4a, left panel) with a 32Å pore in the middle. This region consists of a ring of 2-helix bundles (Fig. 4a, lower right panel) formed by helices α2 and α3 of the TraFCT antenna. These helices each form a ring, with the α3 ring inside the α2 ring, suggesting that α2 is the trans-membrane helix contacting the membrane. Two lines of evidence indicate that this region is inserted in the outer membrane and forms the outer membrane channel. Firstly, α2, the external helix in the 2-helix bundle, has all the features of a trans-membrane helix: it is amphipathic with its hydrophobic side expected to contact the membrane and is strongly predicted to form a trans-membrane helix by TMPred (http://www.ch.embnet.org/software/TMPRED_form.html). Secondly, when a FLAG-tag is inserted between helices α2 and α3 of TraF in the full-length core complex, the tag is found exposed extracellularly (Fig. 4b), demonstrating that the two-helix bundle projects across the outer membrane. Altogether, these results unambiguously assign the 2-helix bundle region of TraF/VirB10 as the channel-forming region of T4S systems.
Figure 4.
Trans-membrane region of the T4S system outer membrane complex and proposed mechanism of pore opening/closure. a. Left panel: surface diagram of complex viewed from the extracellular milieu. The two lines define the hydrophobic central region around the pore. Top right panel: same as at left but showing (in yellow) the ring of Trp residues at the base of α2. Bottom right panel: ribbons diagram of the trans-membrane helices with the internal and external ring of helices colour-coded in green and red, respectively. b. Extracellular detection of the FLAG-tag located between helices α2 and α3 of TraF. A FLAG-tag was introduced in the loop between the α2 and α3 helices of TraF as described in methods. Upper and lower left panels: control sample expressing the wild-type T4SS core complex. Upper and lower right panels: cells expressing the FLAG-tagged T4SS core complex. Differential interference contrast images are shown in the upper panels and the corresponding fluorescence images in the lower panels. c. Proposed mechanism for pore opening and closure. The cryo-EM core complex structure (pale yellow) is superimposed on the crystal structure of the outer membrane complex (pale cyan). The N-terminal lever arm shelf is highlighted in orange and red in the EM and crystal structures, respectively. The position of the lipid on TraN/VirB7 is indicated in black dots. TraN/VirB7 is shown in magenta. Proposed conformational changes are illustrated by arrows. The N-terminal sequences linking the lever arms to sequences in the I-layer are shown in dashed lines colour-coded red and orange for the conformations found in the crystal and EM structures, respectively.
α-helical insertions into outer membranes have been observed only once before, in the case of Wza15,16. In Wza, the C-terminus forms an α-helix and 8 of them in the Wza octamer were shown unambiguously to form the outer membrane channel. The outer membrane channel of the T4S system differs by forming a 2-helix bundle ring system. As in Wza, a tryptophan residue lies at the N-terminal base of the TraF trans-membrane helix contacting the membrane (α2; Fig. 4a, upper right panel)15. This Trp residue is conserved in the vast majority of VirB10 protein family members. In the structure presented here, the outer membrane-inserting region has been cleaved by chymotrypsin (Fig. 1a) and the region around the cleavage is disordered (residues 322 and 344 could not be traced because of poor electron density). As a result, the trans-membrane α2 helices are one turn shorter than those forming the outer membrane channel of Wza. Another consequence of this structural disorder is that the extent of the opening cannot be precisely defined.
The T4S system outer membrane complex, in addition to being much bigger than previously described outer membrane structures (for example Wza or TolC), is also radically different in shape (Supplementary Fig. 9): while the Wza homo-octamer and the homo-TolC trimer are elongated15,17, the hetero-tetradecameric T4S system outer membrane complex spreads out just under the outer membrane, creating a vast contact area with the inner leaflet of the outer membrane. This contact area is mostly mediated by the TraOCT-TraN complex ring. In effect, the TraOCT-TraN complex ring appears to buttress the entire T4S system against the inner leaflet of the outer membrane, possibly allowing the system to exert much greater mechanical forces to extrude substrates than would otherwise be feasible in systems like Wza or TolC which make limited contacts with the inner leaflet of the outer membrane.
Conformational changes in T4S systems
The cryo-EM structure of the T4S system core complex revealed a double-walled structure in the cap of the O-layer (Supplementary Fig. 10a)12. The crystal structure of the cap is not double-walled. This is because the lipid moiety of TraN/VirB7 was not built (the SAD-derived map showed electron density connecting to Cys15 of TraN/VirB7 (the lipidation site) but the density was not sufficiently well-defined for unambiguous fitting). However, when the crystal structure of the outer membrane complex is superimposed on the cryo-EM structure of the entire complex (Fig. 4c), Cys15 of TraN aligns perfectly at the base of the outer wall of the cap, suggesting that this part of the cap might indeed contain the lipidated part of TraN (indicated in Fig. 4c in black dots).
The superposition of the structure of the outer membrane complex and that of the core complex (Fig. 4c; such a superposition is valid as one complex is derived from the other by proteolysis) also reveals that the trans-membrane helices overlap partially with the first half of the inner wall of the cap, suggesting that they might form this region of the cap. However, the crystal structure captures them in a different conformation than the one observed in the cryo-EM structure: while in the cryo-EM structure, they form a narrow vertical constriction, in the crystal structure, they lie in a “relaxed” state at a 45° angle. Thus, the two structures may represent different states. Presumably, by removing the entire N-terminal half of TraF, the constraints that this half places on the N-terminal lever arm of TraFCT might have been removed, releasing the lever arms and leading to relaxation of the trans-membrane helices. Indeed, as illustrated in Fig. 4c, sequences N-terminal to the αn1 helix would directly connect to the I-layer (schematically shown in Fig. 4c by dashed red lines) and are likely to bring the N-terminal arm of each TraF/VirB10 subunit down. This is consistent with the fact that the shelf formed by the N-terminal lever arms of TraF/VirB10 subunits has shifted up in the crystal structure compared to its position in the cryo-EM structure of the full-length core complex (Fig. 4c, Supplementary Figs. 10a and 10b in orange and red, respectively). Thus, we propose that the N-terminal arms of the TraF/VirB10 subunits might act in concert to exert conformational changes in the T4S system channel upon signals sensed by the N-terminal domain of the protein (see below).
In A. tumefaciens, VirB10 is known to undergo a conformational change induced by the energizing T4S system components18. The structure of the T4S system outer membrane complex reveals that VirB10 forms the outer membrane channel. But VirB10 is also known to insert in the inner membrane, making contact not only with the inner membrane channel component VirB8, but also with the ATPases13,19,20. Thus, VirB10 is in a unique position to relay conformational changes taking place in the ATPases and to effect pore opening and closure at the outer membrane. Also, in A. tumefaciens, VirB10 does not directly contact the T-DNA (the T4S substrate), but regulates its handover from the VirB6/VirB8 complex in the inner membrane to VirB9 and VirB2, the major pilin21. As VirB10 lines the interior of the outer membrane complex, it is difficult to envisage how the substrate could not interact with it, unless it is insulated from the substrate by another layer of protein, presumably made of the VirB2 pilin. We thus propose that the VirB2 pilin forms a cylindrical conduit encased within the VirB10 ring. This hypothesis is also consistent with the observation that, in the state captured in the crystal structure (where VirB2 is absent), the TraF/VirB10 trans-membrane helices have somewhat caved in.
The crystal structure of the T4S system outer membrane complex reveals an outer membrane structure of unprecedented size and complexity. This structure is held together by a dense network of protein-protein interactions, which provides a rich targeting ground for inhibitor design. Most striking among them is the extensive interaction that the N-terminal lever arms of the TraFCT subunits make with numerous subunits along the tetradecameric structure. We hypothesized that these sequences are at the heart of a nano-device regulating T4S. If confirmed, this mechanism could become key to the design of inhibitor compounds specifically targeting the T4S machinery.
METHODS SUMMARY
Purification, crystallization, X-ray diffraction data collection
The T4S system core complex was expressed and purified as described previously12. After addition of chymotrypsin, the T4S system outer membrane complex was purified by gel filtration. Crystals were grown by hanging-drop vapour diffusion. Native and single wavelength anomalous diffraction (SAD) data at the selenium edge were collected at ESRF beamline ID14.4 and Soleil's beamline Proxima1, respectively, and processed using the XDS package.
Structure determination and refinement
The structure was solved by molecular replacement using the native dataset and the known 20Å resolution cryo-electron microscopy (cryo-EM) map of the trypsin-cleaved core complex as search model12. 14-fold non-crystallographic symmetry (NCS) averaging and phase extension to 2.8Å resolution yielded a readily interpretable electron density map. This map was used to locate Se atoms using the SAD data set. These heavy metal sites were used to generate SAD phases to 2.6Å resolution, also yielding an interpretable electron density map.
Extracellular localization of a FLAG-tag inserted between the α2 and α3 helices of TraF
A FLAG-tag was introduced at position 332 in the loop between the α2 and α3 helices of TraF. Cells expressing either the wild-type core complex or the FLAG-tagged core complex were grown, fixed, and incubated with anti-FLAG antibodies followed by goat anti mouse IgG1 Texas Red antibodies. Fluorescence was monitored using a Zeiss Axioskop microscope, and images collected using a Hamamatsu Orca ER camera.
Methods
Outer membrane complex purification
Expression of the T4S system core complex from the IBA3c:traN-traFC-ST plasmid was induced using 200μg/l of anhydrotetracyclin12. After incubation overnight at 16°C, the complex was extracted from the membrane fraction and purified using a Strep-Tactin sepharose affinity column (IBA) as described previously12. The elution fractions containing the core complex were pooled together and subjected to limited proteolysis with 2 mg/ml of chymotrypsin for 3 hours at room temperature. The proteolysed sample was then concentrated using a 100kDa cut-off spin concentrator (Amicon) and loaded onto a Superose 6 GL 10/300 (GE Healthcare) gel filtration column in 50 mM TrisHCl pH 8.0, 200 mM NaCl, and 10 mM LDAO. The outer membrane complex eluted as a single peak (Supplementary Fig. 1a).
Selenomethionine labelling
The IBA3c:traN-traFC-ST plasmid12 was transformed into B834(DE3) competent cells (Novagen). Expression of the Selenomethionine (SeMet)-labelled proteins was performed at 16°C overnight in M9 minimal medium supplemented with 50 mg/L of SeMet. Purification of the SeMet-labelled outer membrane complex was as described above for the native complex.
Crystallization and data collection
Large rod-like crystals were grown by vapour diffusion method at 20°C using hanging drops containing 15 mg/ml of the purified native and SeMet-labelled complexes and 20-40% w/v MPD, 100 mM Bis-Tris pH 6.5-7.5. The crystals were flash frozen in liquid nitrogen using the mother liquor as the cryoprotectant. For the SeMet crystals, the oxidation protocol described in Sharff et al. (2000)22 was employed before crystallisation. The oxidised SeMet crystals were further flash-soaked in 0.1% H2O2 in 50% w/v MPD, 100mM Bis-Tris, pH 7.0 before flash freezing for data collection. Data collection was carried out at the beamline ID14-4 at ESRF (Grenoble, France) for the native dataset and at beamline PROXIMA1 at the Soleil Synchrotron (Gif sur Yvette, France) for the oxidised SeMet SAD dataset. Data reduction was carried out using the XDS package23. Data collection statistics are presented in Supplementary Table 1.
Structure determination
The structure was solved by molecular replacement using the native dataset and the known 20Å resolution cryo-electron microscopy (cryo-EM) map of the trypsin-cleaved core complex as search model (Supplementary Fig. 1b)12. The orientation and position of the particle was determined using AMoRe24. The 35 to 20Å resolution range was used and yielded an MR solution with a correlation coefficient of 0.39. 14-fold non-crystallographic symmetry (NCS) averaging with RAVE, MAMA and other programs of the Uppsala suite25-27, in combination with the CCP4 suite28 was used for phase extension to 2.8 Å resolution using the native dataset. This yielded a readily interpretable map with well-defined side chains (Supplementary Fig. 1c, Supplementary Table 1). The three component proteins TraFCT, TraOCT and TraN were traced manually with the program Coot29. Subsequently, this structure was used to locate the Selenium atoms in a SAD data set collected to 2.6Å resolution. Refinement of these sites and phasing (PHASER30), followed by density modification exploiting the 14 fold NCS in PARROT28, yielded a readily interpretable electron density map. After initial tracing by BUCCANEER28, the partial model was used for subsequent cycles of PHASER, PARROT and BUCCANNEER. Further restrained refinement with REFMAC31 and PHENIX 32,33 was carried out using tight restraints on non-crystallographic symmetry throughout the refinement. Final stages of refinement with the map from the SAD dataset showed a density for a modified cysteine in the lipoprotein TraN with small disordered lipid chains attached. However, as the density was not good enough for unambiguous building, we refrained from modeling this modification in the final model. The final model has 94.8% residues in the most favoured region, 4.7% residues in the additionally allowed regions and 0.5% residues in the disallowed region of the Ramachandran plot. Refinement statistics are presented in the Supplementary Table 1.
Extracellular localization of a FLAG-tag inserted between the α2 and α3 helices of TraF
The FLAG epitope was introduced by PCR at position 332 of TraF/VirB10 in the IBA3c:traN-traFC-ST plasmid12, yielding IBA3c:traN-traFFLAG,C-ST. The primer sequences were: 5′-GACGACGACAAGATTCAGTACAACAGCACAGAA-3′ (forward) and 5′-GTCCTTGTAGTCGTTATTACTCTGCGTCTGGTT-3′ (reverse). Expression and purification of the resulting FLAG-tagged T4S system core complex yielded a complex very similar in molecular weight to the wild-type complex (1.05 MDa), indicating that the FLAG-tag does not disturb core complex assembly. E. coli TOP10 cells containing either the IBA3c:traN-traFC-ST plasmid (the expression of which results in the production of the wild-type T4S system core complex)12 or the IBA3c:traN-traFFLAG,C-ST plasmid (the expression of which results in the production of the T4S system core complex containing a FLAG-tag at position 332 of TraF) were grown at 37°C to an OD600nm of 0.6. Protein expression was induced by addition of 200 μg/l of anhydrotetracyclin and cells were incubated at 16°C overnight. Cells were collected by centrifugation, resuspended in ice-cold 4% formaldehyde and incubated on ice for 15 min. The fixed cells were washed twice with PBS and resuspended in PBS. The cell suspensions were applied to poly-L-lysine-treated microscope slides and incubated at room temperature for 20 min. The slides were rinsed in PBS and blocked in 1% goat serum-PBS for 45 min. The primary antibody (anti-FLAG M2 Mab (Sigma), 1:1000 dilution) was applied and incubated on the slide at 4°C overnight. The slides were washed twice with PBS at room temperature and the secondary antibody (goat anti mouse IgG1 Texas Red (Southern Biotech), 1:500 dilution) was applied and incubated on the slide for 1 hour. The slides were then washed twice with PBS and were mounted using DAKO fluorescent mounting medium. The fluorescence microscopy was performed on a Zeiss Axioskop microscope, objective 100x with oil. The images were collected using a Hamamatsu Orca ER camera.
Supplementary Material
Acknowledgments
This work was funded by Wellcome Trust grant 082227 to GW. We thank Dr. Andrew Thompson and the staff of beamline Proxima1 at Soleil, the staff of beamline ID14.4 at ESRF, and Helen Saibil, Elena Orlova and Peter Christie for comments on the manuscript. We thank Dr. Anoop Kumar for help in implementing the immuno-fluorescence experiments.
Footnotes
Declaration of any competing interests. The authors have no competing interests.
Accession numbers. Structure factors and coordinates have been deposited to the PDB (entry codes 3JQO for coordinates and structure factors).
References
- 1.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10:1573–1581. doi: 10.1111/j.1462-5822.2008.01156.x. [DOI] [PubMed] [Google Scholar]
- 5.Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 6.McCullen CA, Binns AN. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol. 2006;22:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 7.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 8.Thomas CM, Nielsen KM. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat Rev Micro. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 9.Fronzes R, Remaut H, Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008;27:2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomis-Ruth FX, Coll M. Cut and move: protein machinery for DNA processing in bacterial conjugation. Curr Opin Struct Biol. 2006;16:744–752. doi: 10.1016/j.sbi.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Bayliss R, et al. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc Natl Acad Sci U S A. 2007;104:1673–1678. doi: 10.1073/pnas.0609535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakubowski SJ, et al. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terradot L, et al. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong C, et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng G, Fronzes R, Chandran V, Remaut H, Waksman G. Protein oligomerization in the bacterial outer membrane (Review) Mol Membr Biol. 2009;26:136–145. doi: 10.1080/09687680802712422. [DOI] [PubMed] [Google Scholar]
- 17.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 18.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci U S A. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods section
- 22.Sharff AJ, Koronakis E, Luisi B, Koronakis V. Oxidation of selenomethionine: some MADness in the method! Acta Crystallogr D Biological Crystallography. 2000;56:785–788. doi: 10.1107/s090744490000370x. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of applied crystallography. 1993;26:795–800. [Google Scholar]
- 24.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallographica Section A: Foundations of Crystallography. 1994;50:157–163. [Google Scholar]
- 25.Jones TA. A set of averaging programs. In "Molecular Replacement". CCP4 Proceedings. 1992:91–95. [Google Scholar]
- 26.Kjeldgaard M, Jones TA. Halloween … masks and bones. In “From First Map to Final Model”. CCP4 Proceedings. 1994:59–66. [Google Scholar]
- 27.Kleywegt G, Jones T. Software for handling macromolecular envelopes. Acta Crystallographica Section D: Biological Crystallography. 1999;55:941–944. doi: 10.1107/s0907444999001031. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.McCoy A, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D: Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Afonine PV, Grosse-Kunstleve RW, Adams PD, The Phenix refinement framework. CCP4 Newsletter. 2005;42 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.