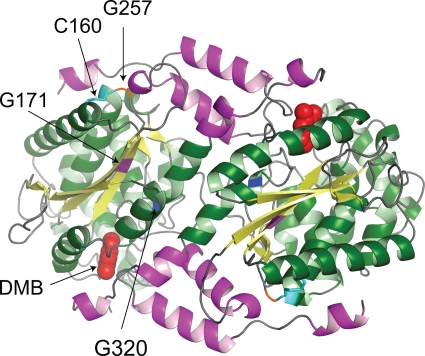
FIG. 3.
Crystal structure model of wild-type CobT protein from S. enterica. Shown is the biologically active dimeric structure of the S. enterica CobT protein in complex with DMB (Research Collaboratory for Structural Bioinformatics Protein Data Bank 1d0s). DMB is shown as space filled in the active sites located at the subunit interface. The arrows indicate the locations of residues affected by class M mutations (G171, G257, and G320). Also shown is the location of residue C160, which forms a disulfide bond with residue C256.