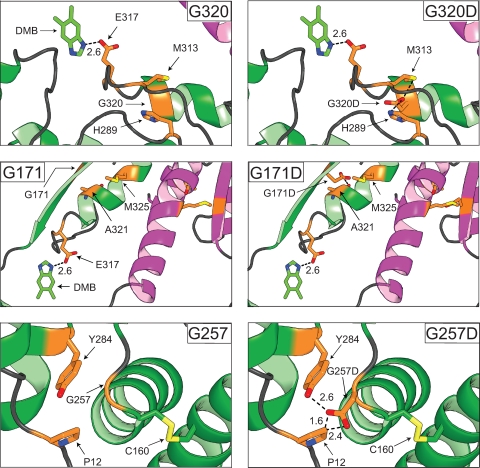
FIG. 4.
Modeling of class M CobT variant proteins. The MacPymol 0.99rev8 software package was used to model in the mutations of interest. As a comparison, the CobTWT protein is located in the left panels while the variant proteins are located in the right panels. One subunit of the proteins is magenta, and the neighboring subunit is green. The variant residues are yellow, and any residues they interact with are green. Interactions between the proposed catalytic residue (E317) and DMB are shown to illustrate how class M substitutions may affect CobT activity. The disulfide bond formed between residues C160 and C256 is yellow. Oxygen and nitrogen atoms in side chains are orange and blue, respectively. Distances are reported in angstroms.