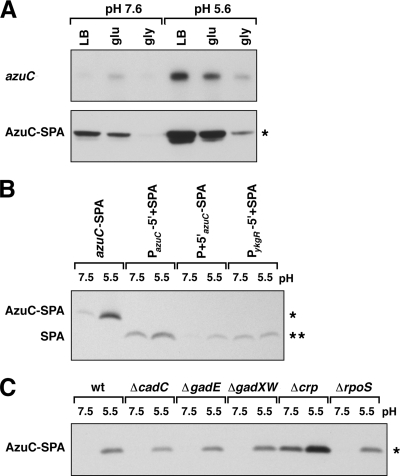
FIG. 5.
Acid induction of azuC. (A) azuC mRNA (top) and AzuC-SPA protein (bottom) levels in MG1655 in LB, minimal glucose (glu), and minimal glycerol (gly) medium buffered at pH 7.6 or 5.6. The RNA band runs at ∼400 nucleotides, consistent with the expected size of the azuC-SPA transcript. Cells were diluted into the respective medium from overnight cultures grown in LB. (B) Acid induction of azuC transcriptional and translational fusions. Extracts from strains containing the SPA-tagged azuC allele (azuC-SPA), a transcriptional fusion to the azuC promoter (PazuC-5′+SPA), a translational fusion to the azuC promoter and 5′ UTR (P + 5′azuC-SPA), or a control transcriptional fusion to the ykgR promoter (PykgR-5′+SPA) were probed for SPA expression in LB-MOPS (pH 7.5) and LB-MES (pH 5.5). In the transcriptional fusions, the azuC and ykgR 5′-UTRs were replaced by the MCS 5′-UTR from pBAD24, and the ORFs were replaced by the SPA tag. In the translational fusion, the azuC ORF was replaced just by the SPA tag (see Materials and Methods). Cultures grown overnight in LB were diluted into 10 ml of LB-MOPS (pH 7.6) or LB-MES (pH 5.6), and cells were harvested at an OD600 of 0.45 to 0.6. (C) AzuC-SPA expression in wild-type and mutant cells grown in neutral and acidic media. Cultures grown overnight in LB were diluted into 5 ml LB-MOPS (pH 7.6) or LB-MES (pH 5.6), and cells were harvested at an OD600 of 0.45 to 0.6. Northern analysis was performed as described for Fig. 3. Western blot analysis was performed as stated for Fig. 2. A single asterisk denotes a band corresponding to the full-length SPA-tagged AzuC protein, and a double asterisk denotes a band corresponding to the SPA peptide.