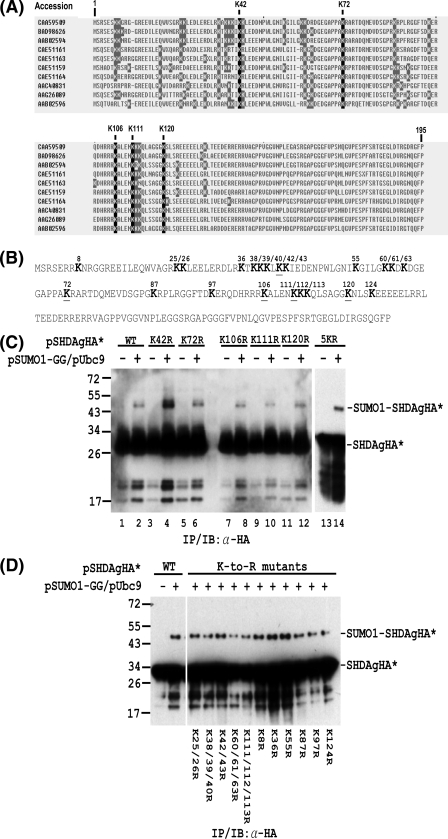
FIG. 5.
Mapping of lysine residues in S-HDAg that are conjugated by SUMO1. (A) Alignment of S-HDAg amino acid sequences from different HDV isolates. The nonconserved and conserved lysine (K) residues are shaded in gray and black, respectively. (B) Amino acid sequence of the Italian HDV isolate used in the in vivo sumoylation assay. The lysine (K) residues are shown in bold. The underlined K residues denote the lysine residues which are conserved among different HDV isolates. (C) The SUMO acceptor lysines within S-HDAg are not limited to the five conserved lysines. In Huh-7 cells, expression plasmids encoding wild-type S-HDAgHA or its mutants containing K-to-R substitutions at the conserved lysine(s) were transfected with or without the SUMO1-GG and Ubc9 expression plasmids. The whole-cell extracts were then prepared and subjected to IP/IB with an antibody to the HA tag. (D) Multiple lysine residues are SUMO1 acceptors within S-HDAg. Expression plasmids encoding wild-type S-HDAgHA or its mutants containing K-to-R substitution(s) were used for in vivo sumoylation assay as in panel C. The positions of the S-HDAgHA species (denoted as SHDAgHA*) and the SUMO1-conjugated form (denoted as SUMO1-SHDAgHA*) are marked on the right. Expression plasmids encoding wild-type S-HDAgHA or its mutants are indicated at the top or bottom.