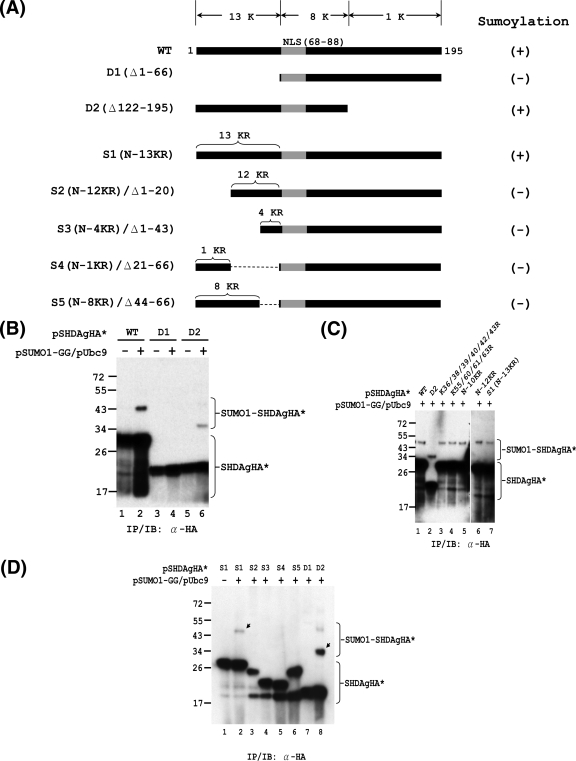
FIG. 6.
Mapping of functional domain required for SUMO modification of S-HDAg. In vivo sumoylation assays were performed with S-HDAg and its mutants. (A) Schematic representation of S-HDAg and its deletion and K-to-R substitution derivatives. The total numbers of lysine (K) residues within the various regions of S-HDAg are indicated at the top. The nuclear localization signal (NLS; amino acids 66 to 88) is shown in gray. The numbers of K-to-R substitutions within the N-terminal region of S1 and its derivatives are indicated on top of these constructs. The ability of the S-HDAg derivatives to undergo sumoylation is summarized on the right. (B) The N-terminally truncated form of S-HDAg loses its ability to be sumoylated. Expression plasmids encoding the wild-type S-HDAgHA (WT) or its deletion mutants, D1 and D2, were used for in vivo sumoylation and subsequent IP/IB assay with anti-HA antibody. (C) The N-terminal 13 lysines are not the exclusive SUMO acceptor lysines of S-HDAg. Expression plasmids encoding wild-type S-HDAgHA and its derivatives (as indicated) were used for in vivo sumoylation and subsequent IP/IB assay with anti-HA antibody. (D) The N-terminal 66 aa of S-HDAg are required for its SUMO modification. The indicated expression plasmids were used for in vivo sumoylation assay as mentioned above. The arrows indicate SUMO1-modified S-HDAgHA* species.