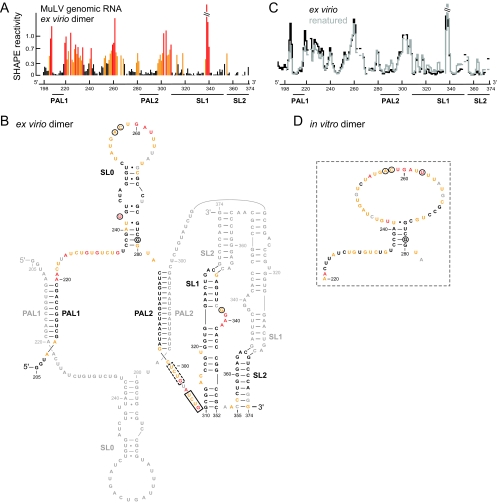
FIG. 3.
SHAPE reactivities and secondary structure model for the ex virio MuLV genomic RNA dimer. (A) Histogram of SHAPE reactivities for authentic ex virio MuLV genomic RNA isolated from virions. (B) Secondary structure model for the ex virio dimer. Nucleotides are colored according to their reactivity as shown in panel A. For clarity, only one of the two dimer strands is annotated in color; a small number of nucleotides in the colored strand (in gray) were not analyzed due to high background. Nucleotide differences between MuLV and MuSV are circled. The nucleocapsid binding site characterized by NMR (13) plus a similar site accessible in the SHAPE-constrained secondary structure are highlighted in solid and dashed boxes, respectively. (C) Comparison of the SHAPE profiles for the ex virio RNA with that of the same RNA that has been heated (to 90°C) and renatured in vitro. The y-axis scale is the same as that used in panel A. (D) Structure model for the region of the in vitro dimer that corresponds to the SL0 motif in the ex virio RNA.