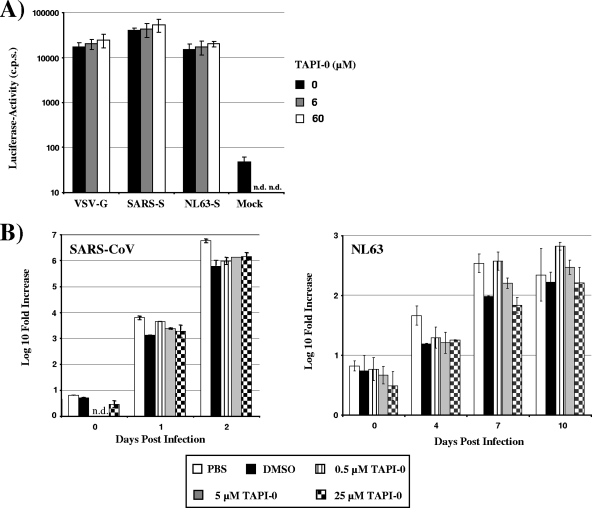
FIG. 3.
Shedding of ACE2 is dispensable for SARS-CoV and NL63 spread. (A) In order to determine the importance of TACE activity for SARS-S- and NL63-S-driven infectious entry, ACE2-transfected 293T cells were incubated with the indicated concentrations of TAPI-0 and infected with infectivity-normalized lentiviral pseudotypes bearing the indicated glycoproteins. At 72 h postinfection, the cells were lysed and the luciferase activities in cell lysates were determined by employing a commercially available kit. (B) To assess the importance of ACE2 shedding to SARS-CoV and NL63 spread, Vero E6 cells were pretreated with the indicated concentrations of TAPI-0 or pretreated with dimethyl sulfoxide (DMSO) as a control and then infected with SARS-CoV (Frankfurt strain) or NL63 at an MOI of 0.001. Supernatants of the infected and noninfected cells were taken at the indicated time points postinfection, and the number of viral genome copies was determined by real-time reverse transcription-PCR (RT-PCR). The following primers and probes were used for detection of the SARS-CoV genome: BNITMSARS1 (5′-TTATCACCCGCGAAGAAGCT-3′) (forward primer), BNITMSARAs2 (5′-CTCTAGTTGCATGACAGCCCTC-3′) (reverse primer), BNITMSARP (5′-FAM-TCGTGCGTGGATTGGCTTTGATGT-TAMRA-3′) (probe). For detection of the NL63 genome, the following primers and probes were used: 63RF2 (5′-CTTCTGGTGACGCTAGTACAGCTTAT-3′) (forward primer), 63RR2 (5′-AGACGTCGTTGTAGATCCCTAACAT-3′) (reverse primer), and 63RP (5′-FAM-CAGGTTGCTTAGTGTCCCATCAGATTCAT-3′-TAMRA) (probe). The SARS-CoV and NL63-specific primers both recognize ORF1B sequences. The result of a representative experiment carried out in duplicates is shown.