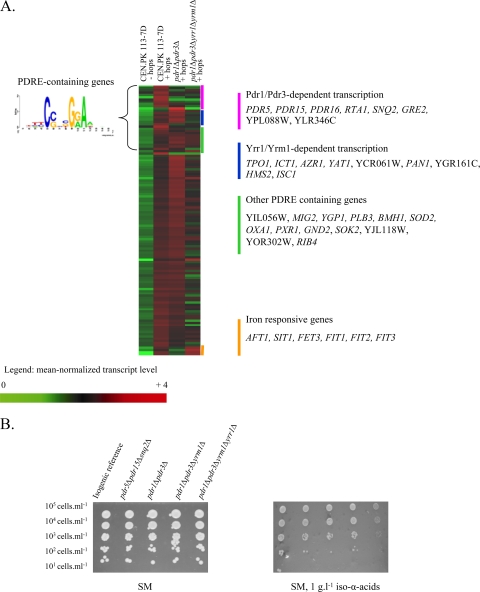
FIG. 3.
Involvement of the PDRE regulatory network in hop iso-α-acid tolerance. (A) Heat map and identities of genes with a hop iso-α-acid-responsive transcription profile. Transcript levels were determined with Affymetrix Genechips YG-98 and are represented as the average of two or three independent experiments. Shown are the mean normalized transcript levels of 120 genes found to be upregulated when comparing the presence of 0.2 or 1 g liter−1 hop iso-α-acids to the reference condition in CEN.PK113-7D. The transcript levels of these genes in the pdr1Δ pdr3Δ and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains grown in the presence of hop iso-α-acids are also indicated. (B) Phenotypic analysis of PDR-deficient mutants: the isogenic reference strain and the pdr5Δ pdr15Δ snq2Δ, pdr1Δ pdr3Δ, pdr1Δ pdr3Δ yrr1Δ, and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains were spotted at 105 cells ml−1 (and three 10-fold serial dilutions) on synthetic agar medium (SM) in the presence or absence of 1 g liter−1 hop iso-α-acids under aerobic conditions (similar results were obtained under anaerobic conditions [data not shown]).