Abstract
Copper is an important cofactor for many enzymes; however, high levels of copper are toxic. Therefore, bacteria must ensure there is sufficient copper for use as a cofactor but, more importantly, must limit free intracellular levels to prevent toxicity. In this study, we have used DNA microarray to identify Staphylococcus aureus copper-responsive genes. Transcriptional profiling of S. aureus SH1000 grown in excess copper identified a number of genes which fall into four groups, suggesting that S. aureus has four main mechanisms for adapting to high levels of environmental copper, as follows: (i) induction of direct copper homeostasis mechanisms; (ii) increased oxidative stress resistance; (iii) expression of the misfolded protein response; and (iv) repression of a number of transporters and global regulators such as Agr and Sae. Our experimental data confirm that resistance to oxidative stress and particularly to H2O2 scavenging is an important S. aureus copper resistance mechanism. Our previous studies have demonstrated that Eap and Emp proteins, which are positively regulated by Agr and Sae, are required for biofilm formation under low-iron growth conditions. Our transcriptional analysis has confirmed that sae, agr, and eap are repressed under high-copper conditions and that biofilm formation is indeed repressed under high-copper conditions. Therefore, our results may provide an explanation for how copper films can prevent biofilm formation on catheters.
Staphylococcus aureus is responsible for a diverse range of infections (12) and continues to be a significant health care problem worldwide. To be a successful pathogen, S. aureus must sense and respond to changing environmental stresses. A key environmental stress encountered during infection is the varying concentration of the transition metal, copper. Copper is an important cofactor for many enzymes, including those essential for catabolic pathways and energy metabolism; however, high levels of copper are toxic (30, 39). It has been proposed that toxicity can occur through copper catalyzing the generation of superoxide or other reactive oxygen species (ROS), leading to cell damage and death; binding to free thiol groups, affecting the function/folding of proteins; or competing with other metals for binding sites, leading to their displacement and enzyme inactivation (41). Therefore, bacteria must ensure that there is sufficient copper for use as a cofactor but, more importantly, must limit free intracellular levels to prevent toxicity.
Studies have shown that in a host environment, while free copper levels are very low, especially in the blood, copper concentrations are higher in some tissues like the lungs (3) and have been shown to increase during infection and inflammation (1). Therefore, it is possible that S. aureus and other bacteria will need to respond to increased copper levels in a host. This is supported by the fact that copper resistance mechanisms are important for virulence, as shown in Listeria monocytogenes and Pseudomonas aeruginosa (10, 33.), and transcription of the Mycobacterium tuberculosis copper-exporting P1-type ATPase gene is induced in mouse lung tissue and tubercle bacillus-infected macrophages (13, 40).
Bacteria have developed a number of mechanisms for dealing with excess copper. These mechanisms involve the efflux and sequestration of copper and a global adaptive response which can involve induction of other stress regulons (18, 41, 47). However, the mechanisms used and particularly the coordination of regulatory responses can vary significantly between species. Efflux mechanisms include the ubiquitous CopA/CopB P1-type ATPase transporters (44) and copper binding proteins, such as the CopZ family, which chaperone the copper ions intracellularly for incorporation/use by other copper binding proteins such as the efflux transporters (29). Most gram-negative bacteria also encode a multicopper oxidase (MCO), which is required for the periplasmic oxidation of Cu(I) to Cu(II) following P1-type ATPase transport of Cu(I) from the cytoplasm (31). Five types of bacterial copper-responsive regulators have been identified to date. CopY is a copper-responsive repressor in Enterococcus and Streptococcus spp. (29). In Escherichia coli, CueR is a MerR homologue and CusRS is a two-component regulator system, both of which activate transcription in response to excess copper (18), whereas CsoR is a copper-responsive repressor in M. tuberculosis (20) and Bacillus subtilis (38).
All sequenced S. aureus strains have an operon encoding a P1-type ATPase (copA) and a copper binding chaperone homologue (copZ) (37). The expression of the copAZ operon is induced by copper, and mutational analysis has shown that CopA transports copper from the cell and hence is required for S. aureus tolerance of excess copper (37). The identification of copA by signature-tagged mutagenesis in a murine renal abscess model (21), and the induction of copA expression following phagocytosis by human neutrophils (46) suggest that CopA is important for S. aureus virulence. A second putative copper-transporting P1-type ATPase (copB) and a multicopper oxidase (mco) have also been identified in a small number of S. aureus strains, and mco has been shown to have a role in copper and oxidative stress resistance (36). It is thought that the copB and mco genes are not essential for normal staphylococcal growth, as they are not found in all S. aureus strains. To date it is not known what other mechanisms are involved in S. aureus copper homeostasis, how gene expression is regulated in response to copper, and what roles these mechanisms have in S. aureus virulence. It is also has not been established how copper toxicity occurs in S. aureus or other gram-positive bacteria.
In this study, we have used DNA microarray to identify S. aureus copper-responsive genes. Transcriptional profiling of S. aureus SH1000 grown in excess copper identified a number of genes which fall into four groups, suggesting that S. aureus has four main mechanisms for adapting to high levels of environmental copper, including induction of the oxidative stress response. Our phenotypic analysis demonstrates that resistance to oxidative stress and particularly H2O2 scavenging are important for S. aureus growth in high levels of copper. Our microarray results also showed that copper represses two major global virulence gene regulators, Sae and Agr. This has important implications for the effect of copper on virulence and the antimicrobial uses of copper. Our previous studies have demonstrated that Eap and Emp proteins, which are positively regulated by Agr and Sae, are required for biofilm formation under low-iron growth conditions (16). Biofilm formation is an important S. aureus virulence factor, as S. aureus is a major pathogen associated with nosocomial infections due to its ability to colonize a wide range of medically implanted devices, which are colonized as biofilms. Copper films on catheters have been shown to prevent biofilm formation, but the underlying mechanisms involved were unknown (23). Thus, this study shows that excess copper represses the expression of positive regulators of biofilm formation sae and agr and a biofilm factor, eap, and hence represses S. aureus biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table 1 and were stored at −80°C in 10% (vol/vol) glycerol TSB (Trypticase soy broth; BBL). S. aureus strains were plated out fresh from frozen stocks onto 6% defibrinated horse blood agar for each biofilm assay. Strains for biofilm assays and copper growth assays were cultured under iron-restricted conditions in CRPMI, RPMI 1640 tissue culture medium (Sigma Ltd) which had been depleted of iron by batch incubation with 6% (wt/vol) Chelex 100 (Sigma Ltd) and then supplemented with 10% RPMI 1640 to provide trace elements required for growth (26). CRPMI cultures were incubated statically for 24 h at 37°C in 5% CO2 in air. Various concentrations of CuSO4, MnCl2, MgCl2, and CaCl2 were added to achieve metal-replete conditions.
TABLE 1.
Bacterial strains and plasmids used in this study
| S. aureus strain | Description | Source or reference |
|---|---|---|
| Newman | Wild type | 9 |
| SH1000 | NCTC 8325-4 with rsbU mutation repair | 15 |
| SH1000 copA | SH1000 with partial copA deletion | 37 |
| SH1000 ahpC | SH1000 ahpC::tet | 7 |
| SH1000 katA | SH1000 katA::Tn917 | 7 |
| SH1000 ahpC katA | SH1000 ahpC::tet katA::Tn917 | This work |
| SH1000 sodA | SH1000 sodA::Tn917 | 6 |
| SH1000 sodM | SH1000 sodM::tet | 17 |
| SH1000 sodM sodA | SH1000 sodM::tet/sodA::Tn917 | 17 |
Transduction of mutations into S. aureus strain Newman.
The kat::Tn917 mutation was transferred to S. aureus strain SH1000 ahpC::tet by transduction with phage phi11 (5) from SH1000 katA. Transductants were selected on LK agar plates (LK medium consists of 1% tryptone, 0.5% yeast extract, 0.7% KCl, and 1.5% agar) containing 10 μg ml−1 of either kanamycin or tetracycline and 0.05% sodium citrate. Colonies containing the relevant mutation were confirmed by PCR using primers katF and katR (Table 2).
TABLE 2.
Primers used in this study
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| agrC | agrC-F | CATATTCGCAAATTGATGCG |
| agrC-B | CCTAAACCACGACCTTCACC | |
| sodM | sodM-F | CAATGGAGTTTCATCACGACA |
| sodM-B | GTAGGCATGCTCCCAAACAT | |
| copA | copA-F | ATATGACTTGTGCCGCATGTT |
| copA-B | GCTTAATCAATTTATGTTGTAGCGC | |
| copZ | copZ-F | CGGATCCATGTCACAAGAAATTTTAA |
| copZ-B | GAAGCTTTTAAACGACATCGTAACCT | |
| saeS | SaeS-F | TTGTTGCGCGAGTTCATTAG |
| SaeS-F | ACGGAAATTACGCAAGCAAT | |
| ahpF | SAS0357-F | CGGTGCTCATGTCTTCGTAA |
| SAS0357-B | GTGGTGGTCCTGCTAGTGGT | |
| oppA | SAV0990-F | AAACCGGTGTTAGGTGTTGC |
| SAV0990-B | GCTCTATCAGCTGCCGTACC | |
| katA | katF | GACCTCTTTTAATGCAAGATA |
| katR | GAAACTACATATCAAATTTAT | |
| 16S | 16S-F | GATCCTGGGTCAGGATG |
| 16S-R | CTAGAGTTGTCAAAGGATG |
RNA isolation and quantification.
RNA was isolated from three independently grown 20-ml cultures of S. aureus. S. aureus strains were grown by shaking them in defined medium (DM) at 37°C overnight, as described by Townsend and Wilkinson (42), and then diluted 1:200 into fresh DM. Bacteria were then pregrown in DM until mid-log phase (optical density at 600 nm [OD600] = 0.4). A total of 100 μM copper sulfate was then added to create conditions of copper excess, whereas control cultures were grown in the absence of copper. Test and control cultures were then incubated at 37°C with shaking for a further 30 min. Ten milliliters of exponentially growing cultures was pretreated with an equal volume of RNAprotect (Qiagen Inc.) before cell harvesting at 8,000 × g for 15 min. The cell pellet was then resuspended in 1 ml Tri reagent (Sigma), transferred to a centrifuge tube containing 0.1-mm zirconia/silica beads (Biospec Products Inc.), and processed in an FP120 FastPrep cell disruptor (MP Biomedicals, Irvine, CA). BCP (1-bromo-3-chloropropane) was then added to the lysates, which were centrifuged at 16,000 × g for 15 min, and then the RNA was precipitated with 100% ethyl alcohol. RNA was further purified using DNase I and RNeasy minikits (Qiagen), according to the manufacturer's instructions. Following the final purification, RNA was dissolved in RNase-free water, and the concentration was determined by measuring the absorbance at 260 nm. Finally, the purity and integrity of RNA were checked through agarose gel electrophoresis.
Transcriptional profiling.
DNA microarray experiments were performed using three independently cultured sample sets to isolate the RNA, and each set was run independently to generate triplicate data. S. aureus grown in the absence of copper served as the control, whereas S. aureus grown in excess copper as described above served as the experimental set. Labeling of RNA was carried out using 5 μg of RNA, followed by synthesizing of cDNA using reverse transcriptase (Invitrogen), as described previously (27). Briefly, cDNA was generated using random hexamers as primers for reverse transcription. The primers were annealed at 70°C for 10 min, followed by snap-freezing in ice for 1 min to total RNA (5 μg), and were extended with SuperScript II reverse transcriptase (Invitrogen) with 0.1 M dithiothreitol-12.5 mM deoxynucleoside triphosphate-5-(3-aminoallyl)-dUTP mix (Αmbion, Αustin, ΤX) mix at 42°C overnight. Two independent sets were performed. In one experiment, the control sample was labeled with Cy3, and the test sample was labeled with Cy5. For the other experiment, Cy5 was used to label the control sample, and Cy3 was used to label the test sample. Fluorescently labeled cDNA was purified by the Qiagen PCR purification kit. After being dried, the cDNA was dissolved in a hybridization buffer composed of 30% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and a 1:100 volume of sheared salmon sperm DNA and heated at 98°C for 2 min. The denatured cDNA was applied to a S. aureus genome microarray version 5.0 DNA chip (Pathogen Functional Genomic Resource Center—The Institute of Genomics Research [PFGRC-TIGR]) (http://pfgrc.jcvi.org/) under a glass slide, and hybridization was carried out overnight at 42°C. After hybridization, the slides were washed at room temperature in wash buffer I (1× SSC and 0.2% SDS) twice for 10 min, in wash buffer II (0.1× SSC and 0.1% SDS) once for 10 min, and in wash buffer III (0.1× SSC) twice for 10 min. The slides were dried by centrifugation at 500 rpm for 5 min.
Data analysis.
Hybridization signals were scanned using a Axon 4000B scanner with Acuity 4.0 software. Scans were analyzed using TIGR Spotfinder software, and the data set was normalized by applying the LOWESS algorithm using TIGR-MIDAS software (www.tigr.org/software/). Significant changes of gene expression were identified with significance analysis of microarrays (SAM) software using one class mode. SAM assigns a score to each gene on the basis of the change in gene expression relative to the standard deviation of repeated measurements. The “q value” assigned to each gene corresponds to the lowest false discovery rate at which the gene is called significant (43). Three independent cultures were used to prepare RNA.
RNA extraction and Northern blotting.
Exponentially growing cultures in CRPMI were pelleted and resuspended in RNAlater, as directed by the manufacturer (Ambion). Total RNA was then extracted using the hot phenol method, as described by Schmitt et al. (32), but with 300 μg ml−1 lysostaphin added to the initial cell lysis step. A total of 10 μg RNA was electrophoresed on a denaturing 1.5% formaldehyde gel, and the RNA was transferred to a nylon filter by Northern blotting overnight. The filters were hybridized overnight at 65°C in Church Gilbert's buffer (0.5 M Na2HPO4, 0.5 M NaH2PO4, 7% [wt/vol] SDS, 1 mM EDTA) with probes labeled with [α-32P]dCTP (PerkinElmer), using the Klenow fragment of DNA polymerase I and random hexamers as primers. Probes were constructed by PCR using the appropriate primer pairs shown in Table 2. Filters were washed with 3× SSC/0.1% (wt/vol) SDS and exposed to X-ray film at −80°C. The blots were then stripped of probes by washing the filters with 2 to 3 changes of boiling 0.1% (wt/vol) SDS and rinsing with 2× SSC. Transcripts were evaluated using ImageJ 1.41 software (downloaded from http://rsbweb.nih.gov/ij/). Values were normalized against 16S rRNA transcripts and expressed as a percentage of the level of transcription in the wild type.
MIC determination.
The MIC of Cu was calculated as the lowest concentration at which there was no growth after 24 h of exposure. S. aureus was grown overnight in CRPMI at 37°C in 5% CO2 in air, diluted to an optical density at 595 nm of 0.05 in fresh warmed CRPMI, and inoculated into triplicate wells of 96-well flat-bottomed tissue culture plates (Nunc) with differing concentrations of copper chloride. A total of 500 U/ml of catalase and 10 U/ml of superoxide dismutase (SOD) were added when required. The plates were then incubated statically at 37°C in 5% CO2 in air for 24 h. After incubation, the plates were read at 595 nm in a plate reader (Bio-Rad), and the MIC was determined. Grown cultures were blanked with medium containing the equivalent copper concentration. The mean results and standard deviations shown have been calculated from at least three independent experiments. A two-tailed Student's t test was used to determine the differences in growth between mutant and wild-type strains. Differences were considered statistically significant when P was <0.05.
Biofilm formation assay.
Quantitative measurement of staphylococcal adherence to polystyrene, indicative of biofilm formation, was done in a microtiter assay as described previously (8), with the following modifications. Bacteria were grown overnight in CRPMI at 37°C in 5% CO2 in air, diluted to an optical density at 595 nm of 0.1 in fresh CRPMI, and inoculated into quadruple wells of 96-well flat-bottomed tissue culture plates (Nunc). Appropriate concentrations of CuSO4, MnCl2, MgCl2, or CaCl2 were added, and the plates were then incubated statically at 37°C in 5% CO2 in air for 24 h. After incubation, plates were read at 595 nm in a plate reader (Bio-Rad) to monitor growth. Wells were then washed three times with sterile PBS (phosphate-buffered saline) to remove nonadherent cells, dried at 65°C, and stained with 1% (wt/vol) safranine. After being stained, plates were read in a plate reader at 490 nm to determine the extent of biofilm formation. The mean results and standard deviations shown have been calculated from at least four independent experiments. A two-tailed Student's t test was used to determine the differences in S. aureus biofilm formation among the different metal ions. Differences were considered statistically significant when P was <0.05.
RESULTS
Excess environmental copper causes a global adaptive response involving a number of stress responses and virulence gene regulators.
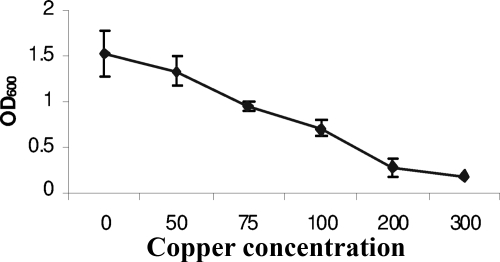
To investigate the S. aureus response to sudden copper stress, microarray analysis was used to determine the transcriptome of exponentially growing S. aureus exposed to excess copper. To determine the appropriate concentration of copper for the transcriptional profiling, S. aureus SH1000 was first grown in chemically defined medium (DM) with the addition of various concentrations of CuSO4 (Fig. 1). S. aureus grew to maximal levels in DM in the absence of added copper, whereas the MIC of CuSO4 in the defined media was 200 μM. Therefore, the subinhibitory concentration of 100 μM CuSO4 was added to DM under the transcriptional profiling excess copper condition. A doubling time of 90 min was observed in the presence of 100 μM CuSO4 compared to that of 60 min in the absence of copper.
FIG. 1.
Graph showing growth of S. aureus SH1000 in DM containing various concentrations of copper sulfate (in μM).
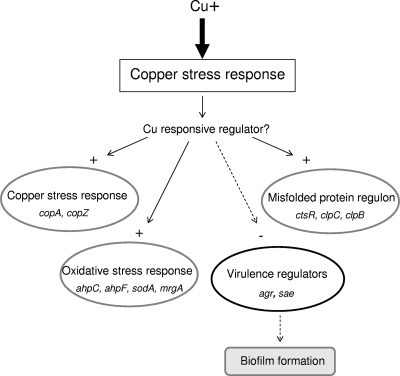
To determine the effect of excess copper on S. aureus gene expression, exponentially growing S. aureus was exposed to excess copper for 30 min as described in previous studies (41). Very little growth occurred in the copper-containing culture after 30 min (the OD600 was from 0.4 to 0.44), whereas some growth occurred in the control culture (the OD600 increased from 0.4 to 0.53). This reflects a 17% inhibition in the growth of the test culture during the 30-min copper exposure time. Total RNA was extracted from exponentially growing SH1000 grown in DM for 30 min with or without the addition of 100 μM CuSO4. cDNA samples were hybridized with the PFGRC S. aureus genomic microarray. After statistical evidence with a ≥2.0-fold difference as the threshold change and a P value of less than 0.05 was applied, microarray analysis showed that exposure to excess copper induced 39 genes and repressed 58 genes which fall into four main groups (see Table S1 in the supplemental material). As shown in Fig. 2, excess copper induces a copper stress response, which includes copper export and chaperone mechanisms and the misfolded protein and oxidative stress responses. The microarray analysis also shows that there is repression of the protein biosynthesis genes, a number of transporters, and the virulence gene global regulators agr and sae. There are also a large number of hypothetical genes which are differentially regulated by excess copper (Tables 3 and 4).
FIG. 2.
Model of the S. aureus adaptive copper response. Excess copper induces the copper, protein misfolding, and oxidative stress regulons, represses virulence gene regulators Agr and Sae, and hence, represses biofilm formation. Arrows indicate positive regulation, and dotted arrows indicate negative regulation.
TABLE 3.
Genes upregulated in excess copper
| Locus tag | Genea | Description | Fold change |
|---|---|---|---|
| Direct copper homeostasis mechanism genes | |||
| SACOL2573 | copZ | Copper ion binding protein | 6.8 |
| SAV2557 | copA | Copper-transporting ATPase | 6.64 |
| SAS2591 | NA | DNA binding protein | 2.24 |
| SAS1326 | phoU | Putative phosphate transport system protein | 2.22 |
| SAS1804 | NA | Putative exported protein | 2.1 |
| Oxidative stress resistance genes | |||
| SAV1713 | NA | Probable thiol peroxidase | 2.77 |
| SAS2042 | mrgA | Putative non-heme-containing ferritin | 2.71 |
| SAS0357 | ahpF | Alkyl hydroperoxide reductase subunit F | 2.68 |
| SAR0135 | sodM | Superoxide dismutase | 2.56 |
| SACOL1155 | trxA | Thioredoxin | 2.42 |
| SAV0764 | trxB | Thioredoxin reductase | 2.20 |
| SAV2187 | NA | Quinine oxidoreductase | 2.14 |
| Misfolded protein response genes | |||
| SACOL0570 | clpC | ClpC ATPase | 3.25 |
| SACOL0833 | clpP | ATP-dependent ClpP protease | 2.58 |
| SAV0975 | clpB | ClpB chaperone | 2.13 |
| SAS0479 | ctsR | CtsR transcriptional repressor | 2.44 |
NA, not applicable.
TABLE 4.
Genes downregulated in excess copper
| Locus tag | Gene | Description | Fold change |
|---|---|---|---|
| Transport genes | |||
| SAV0990 | oppA | Oligopeptide transporter binding protein | −2.16 |
| SAV2316 | potE | Transport protein | −2.13 |
| SAV0950 | mnhC | Putative monovalent cation/H+ antiporter subunit C | −2.1 |
| SAS2358 | opp-1A | Oligopeptide transporter binding protein | −2.09 |
| Regulator genes | |||
| SACOL2024 | agrD | Accessory gene regulator protein D | −2.17 |
| SACOL0765 | saeS | Sensor histidine kinase SaeS | −2.00 |
| SACOL0766 | saeR | Response regulator SaeR | −2.01 |
| Cofactor and carrier biosynthetic genes | |||
| SAV1769 | ribA | Riboflavin biosynthesis | −2.48 |
| SACOL0538 | ispE | 4-Diphosphocytidyl-2C-methyl-d-erythritol kinase | −2.21 |
| SAV1771 | ribD | Riboflavin-specific deaminase | −2.11 |
| SACOL1819 | ribE | Riboflavin synthase, alpha subunit | −2.00 |
| Protein biosynthesis genes | |||
| SACOL0590 | 30S ribosomal protein L7 | −2.09 | |
| SACOL0591 | rpsL | Ribosomal protein S12 | −2.25 |
| SACOL0592 | rpsG | Ribosomal protein S7 | −2.36 |
| SACOL0593 | fusA | Translation elongation factor | −2.29 |
| SACOL2212 | rplQ | Ribosomal protein L17 | −2.17 |
| SACOL2213 | rpoA | DNA-directed RNA polymerase | −2.27 |
| SACOL2216 | rpmJ | Ribosomal protein L36 | −2.75 |
| SACOL2217 | infA | Translation initiation factor IF-1 | −2.18 |
| SACOL2220 | rplO | Ribosomal protein L15 | −2.02 |
| SACOL2224 | rplF | Ribosomal protein L6 | −2.18 |
| SACOL2225 | rpsH | Ribosomal protein S8 | −2.19 |
| SACOL2226 | rpsN2 | Ribosomal protein S14 | −2.33 |
| SACOL2227 | rplE | Ribosomal protein L5 | −2.65 |
| SACOL2228 | rplX | Ribosomal protein L24 | −3.09 |
| SACOL2229 | rplN | Ribosomal protein L2 | −2.14 |
| SAV2247 | rplB | Ribosomal protein L36 | −2.75 |
| SACOL2234 | rplV | Ribosomal protein L22 | −2.09 |
| SACOL2235 | rpsS | Ribosomal protein S19 | −2.17 |
| SACOL2238 | rplD | Ribosomal protein L4 | −2.24 |
| SACOL2239 | rplC | Ribosomal protein L3 | −2.75 |
Excess copper induces copper homeostasis mechanisms.
Our previous studies identified a direct mechanism for copper homeostasis that involved induction of the copAZ operon, which encodes the CopA and CopZ proteins, both of which have metal binding domain motifs MXCXXC (37). The transcriptional profiling experiments showed that both copA and copZ are significantly induced by copper (fold changes of 6.6 and 6.8, respectively), thus demonstrating that the microarray results are representative of the global response to excess copper (36) (Table 3). However, the transcriptional profiling did not identify any other known copper homeostasis homologues or any other proteins with metal binding motifs, although it did identify a number of hypothetical genes with increased expression in excess copper of the same order as that of the copA and copZ genes (see Table S1, MW1746, 5.8-fold; SACOL1858, 4.61-fold; and SAV0454, 4.05-fold, in the supplemental material). These may represent components of novel S. aureus copper homeostasis mechanisms and require further investigation.
The protein translation is downregulated and the misfolded protein and oxidative stress responses are induced by excess copper.
Our transcriptional profiling suggests that an S. aureus adaptive response to copper shock is the temporary downregulation of protein synthesis. The addition of copper to exponentially growing S. aureus resulted in a decrease in the expression of at least 20 genes encoding ribosomal proteins, translation initiation factors, and the RNA polymerase rpoA subunit (Table 4), suggesting that there is a corresponding decrease in protein synthesis upon copper shock.
Copper shock also resulted in the induction of the CtsR-regulated misfolded protein response, as addition of copper induced the expression of the ctsR repressor gene as well as that of the genes encoding the energy-dependent proteolytic complex clpC and clpP and encoding the clpB chaperone (Table 3). In gram-positive bacteria, the Clp proteolytic complex plays an indispensable role in cellular protein quality control by refolding or degrading damaged proteins and is essential for virulence and survival under a number of stress conditions in several pathogenic bacteria, including S. aureus (11). Therefore, these results suggest that sudden exposure to copper damages staphylococcal proteins. Hence, the downregulation of S. aureus protein synthesis could be in direct response to the protein damage caused by the presence of copper.
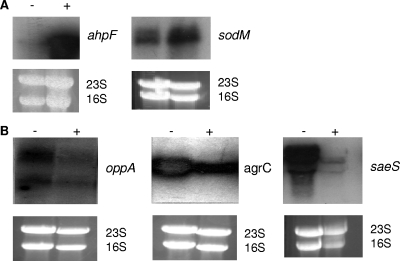
Our microarray analysis also shows that the expression of the oxidative-stress resistance genes alkyl hydroperoxide reductase subunit F (ahpF), superoxide dismutase (sodM), and mrgA is induced in S. aureus more than 2-fold by excess copper (Table 3). S. aureus has a number of mechanisms for protection against oxidative stress. S. aureus MrgA is an important nucleoid protein which demonstrates induced expression during oxidative stress and is important for the compaction of the nucleoid during oxidative stress, which may protect the DNA from damage due to reactive oxygen species (25). SodM is one of two S. aureus superoxide dismutases, SodA and SodM, which catalyze the conversion of the highly reactive O2− to O2 and H2O2 (17). The H2O2 is then reduced to H2O and O2 by the compensatory functions of catalase (katA) and alkyl hydroperoxide reductase (ahpC) (7). AhpF is a homodimeric flavoenzyme carried by the same operon as ahpC, which catalyzes the NADH-dependent reduction of AhpC, therefore regenerating AhpC with every catalytic cycle. Northern blot analysis confirmed the transcriptional profiling results, as ahpF and sodM transcription increased in DM with excess copper (Fig. 3A). This induction of oxidative stress genes agrees with the proposal that copper toxicity is caused partly by Cu1+ catalyzing the formation of highly reactive hydroxyl radicals from H2O2 via the Haber-Weiss reaction (41). However, our microarray study suggests that H2O2 stress is not the only toxic effect of excess copper, as the thioredoxin trxA and thioredoxin reductase trxB genes which are required for combating thiol oxidation are also induced by excess copper. These genes are induced in response to oxidative stress agents that result in increased disulfide bond formation but are not induced by H2O2 (45), suggesting that excess copper also causes thiol oxidation in S. aureus.
FIG. 3.
Northern blot analysis was used to confirm differential regulation of genes in response to excess copper identified by the microarray analysis. S. aureus SH1000 was grown in DM until an OD600 of 0.4 was reached. A total of 100 μM copper was then added, and following 30 min of incubation, total RNA was extracted. Ten micrograms of RNA was resolved by agarose gel electrophoresis and hybridized with radiolabeled ahpF and sodM DNA probes (A) and radiolabeled oppA, agrC, and saeS DNA probes (B). Ethidium bromide-stained 23S and 16S rRNA provided loading controls.
Resistance to oxidative stress is important for S. aureus growth in excess copper.
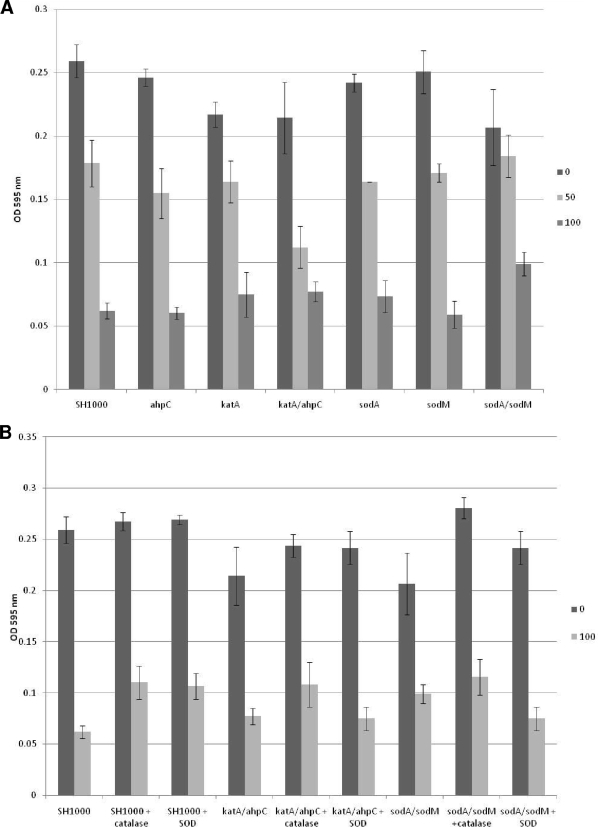
Oxidative stress genes have also been shown to be induced in response to copper stress in P. aeruginosa and M. tuberculosis (41, 47), but the role of the oxidative stress genes in copper resistance has not actually been shown for any bacterium to date. Therefore, to determine the role of the oxidative stress response in S. aureus copper resistance, we investigated the effect of deleting the oxidative stress genes ahpC, katA, sodA, and sodM on S. aureus growth under low- and high-copper conditions. S. aureus SH1000 and the isogenic mutants were grown in CRPMI for 24 h with or without the addition of 50 or 100 μM CuSO4. The copper MIC for SH1000 in CRPMI is 100 μM. This is in contrast to the MIC of 200 μM in the DM used under the microarray conditions, where 100 μM copper only partially inhibited growth (Fig. 1). This is reflective of the increased sensitivity of bacteria to copper grown under nutrient-deprived growth conditions (37).
There was no significant difference between the growth of the wild-type SH1000 and that of the single mutant strains in CRPMI with 100 μM copper, as the growth of all the strains was inhibited (Fig. 4A). There were also no significant differences between the wild-type and single mutant strain growth with 50 μM copper added, as all strains showed a 40 to 50% decrease in growth (Fig. 4A). However, previous publications have shown that AhpC and KatA have compensatory roles, as significant phenotypes were seen only with the double mutant strains (7), and superoxide dismutase (SOD) activity in S. aureus can be provided by both SodA and SodM (17). Therefore, we compared the growth of the double mutant strains SH1000 ahpC katA and SH1000 sodA sodM with wild-type SH1000 in low and high levels of copper. In CRPMI plus 50 μM copper, there was a significant decrease in the growth of SH1000 ahpC katA compared to wild-type SH1000 (P = 0.00002) (Fig. 4A), which suggests that H2O2 scavenging is an important S. aureus copper resistance mechanism.
FIG. 4.
Effect of oxidative stress resistance gene mutations (A) and the addition of exogenous ROS quenchers (B) on growth in excess copper. The S. aureus SH1000 wild type and SH1000 ahpC, katA, ahpC katA, sodM, sodA, and sodM sodA mutant cultures were grown overnight in CRPMI, then diluted to an OD600 of 0.05 in fresh CRPMI with and without 50 or 100 μM copper, inoculated into quadruple wells of 96-well flat-bottomed tissue culture plates, and incubated statically at 37°C for 24 h. After incubation, plates were read at 595 nm to monitor growth. Each point represents the mean result and standard deviation of at least 4 independent experiments. (B) A total of 500 U/ml of catalase or 10 U/ml of superoxide dismutase (SOD) enzymes were added before incubation at 37°C, where indicated.
The importance of H2O2 in S. aureus copper toxicity was further confirmed by the observation that in CRPMI with 100 μM copper, SH1000 sodA sodM mutant growth was significantly increased compared to wild-type SH1000 (P = 0.0002). Therefore, these results confirm the microarray results and show that a proportion of S. aureus copper toxicity is due to oxidative stress and the production of free radicals and that superoxide dismutase activity is actually contributing to copper toxicity. Furthermore, these results show that the oxidative stress mechanisms are required for continued growth in high levels of copper and do not just counteract the initial copper shock.
To further investigate the role of oxidative stress in S. aureus copper toxicity, we compared wild-type SH1000, SH1000 ahpC katA and SH1000 sodA sodM growth under high- and low-copper conditions with and without the addition of exogenous catalase and SOD (Fig. 4B). The addition of external oxidative stress quenchers will determine if the redox reactions are occurring intracellularly only or also at the cell surface. The presence of these ROS quenchers had no effect on growth of S. aureus SH1000 under low-copper conditions. However, S. aureus SH1000 growth was significantly increased when either catalase or SOD was present under high-copper growth conditions (Fig. 4B) (P = 0.03 and 0.01, respectively). Exogenously added catalase or SOD is likely to remain extracellular; therefore, these data suggest that extracellular copper has a toxic effect on S. aureus growth, as exogenous catalase and SOD cannot protect the cell from excess intracellular copper. Addition of catalase to SH1000 ahpC katA also had a protective effect, as there was a significant increase in growth of the mutant in 100 μM copper (P = 0.005). However, exogenous SOD did not increase the survival of the ahpC katA mutant, suggesting that catalase and AhpC are required to convert the H2O2 produced by SOD to H2O and O2. Interestingly, exogenous catalase did not increase SH1000 sodA sodM survival, and in fact, the double mutant was more sensitive to copper in the presence of exogenous SOD (P = 0.04) than in its absence. This suggests that the H2O2 produced by the exogenous SOD is not dealt with correctly in the SH1000 sodA sodM mutant. Therefore, our data suggest that extracellular copper is toxic to S. aureus.
Repression of biofilm formation by excess copper is mediated by the repression of positive biofilm regulators agr and sae.
In addition to induction of the misfolded protein and oxidative stress responses, the microarray analysis also showed that excess copper results in the downregulation of a number of genes, including mnhC, a monovalent cation/H+ antiporter subunit, potE, a putative amino acid transporter, and oligopeptide transporters oppA and opp-1A (Table 4). The repression of oppA in response to excess copper was confirmed by Northern blot analysis (Fig. 3B). The OppBCDFA transporter has been shown to be required for the transport of oligopeptides (14), but the function of Opp-1A is unknown. Opp-1A does have 35% homology with the Escherichia coli NikA nickel transporter (14); therefore, it may have a role in transporting metal ions into the cytoplasm, which may be deleterious when there is excess copper. Hence, its expression is repressed. It is not clear why the expression of these transporters is regulated by copper. The effect of copper on these transport systems may be direct or may be caused by excess copper levels affecting intracellular ion levels per se.
Excess copper also repressed the two-component regulator genes agr and saeRS (Table 4). Agr and Sae are particularly important virulence gene regulators and are essential for S. aureus virulence. The agr system represses the expression of cell wall-associated proteins and induces the expression of several exoproteins during postexponential growth, while the saeRS locus also induces the expression of exoproteins, including α-hemolysin and protein A (2), and is essential for innate immune evasion (46).
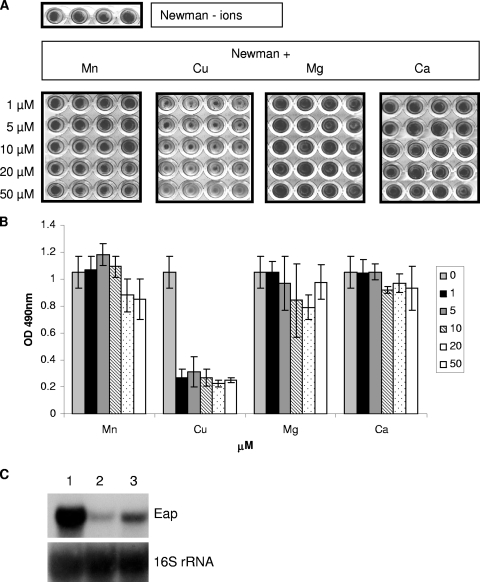
Our previous studies have shown that Agr and SaeRS are required for the induced expression of the Emp and Eap extracellular proteins, which are important for biofilm formation under iron-restricted, nutrient-poor growth conditions (16). Hence, excess copper represses the expression of positive regulators of biofilm formation. Northern blot analysis confirmed the repression of agr and sae expression under excess copper conditions, thus confirming the microarray data (Fig. 3B). Therefore, we investigated the effect of copper as well as other divalent cations on S. aureus strain Newman biofilm formation. Biofilm assays were performed using S. aureus Newman grown in CRPMI with differing concentrations of copper, manganese, magnesium, and calcium (Fig. 5). There was no significant effect on biofilm formation due to the addition of manganese, magnesium, and calcium. However, there was a significant decrease in biofilm formation with even the addition of only 1 μM copper. This was not due to any copper toxicity, as there was no significant difference in S. aureus Newman growth when 1 to 20 μM copper was added, although the addition of 50 μM copper to CRPMI resulted in a 35% decrease in growth (data not shown). Therefore, these results suggest that copper prevents the formation of S. aureus biofilms through the repression of the positive biofilm regulators Agr and Sae. This was further confirmed by Northern analysis, which demonstrated that transcription of the eap gene, which is positively regulated by Agr and Sae and is important for low-iron-induced biofilm formation, is also repressed by excess copper (Fig. 5C).
FIG. 5.
Biofilm (A and B) and Northern blot analysis (C) of S. aureus Newman grown for 24 h in CRPMI, showing the effect of copper and other metal ions on biofilm formation and Eap expression. (B) Results represent the mean and standard deviation of at least four independent experiments. (C) A total of 10 μg of total RNA prepared from S. aureus Newman cells growing exponentially in CRPMI (lane 1), CRPMI with 50 μM Fe2 (SO4)3 (lane 2), or 10 μM copper chloride (lane 3) was resolved by agarose gel electrophoresis and hybridized with eap DNA probe. The blot was then stripped and rehybridized with the 16S rRNA control probe.
DISCUSSION
In this study, we show that the S. aureus response to high levels of copper includes the induction of the copper stress, misfolded protein, and oxidative stress responses, as well as the repression of the global virulence gene regulators agr and sae. These expression studies suggest that copper toxicity in S. aureus is caused by protein damage and oxidative stress through thiol oxidation and the production of ROS. Our phenotypic studies show that the oxidative stress genes are required for S. aureus adaptive growth under high-copper conditions and initial copper shock, therefore demonstrating that oxidative stress has an important role in S. aureus copper toxicity. Furthermore, we show that H2O2 scavenging is a particularly important stress response and that extracellular copper toxicity occurs in S. aureus. These studies also show that the repression of agr and sae leads to the repression of proteins required for the important S. aureus virulence factor, biofilm formation.
Our microarray studies show that protein damage is an important feature of copper stress in S. aureus, as sudden exposure to copper results in the downregulation of S. aureus protein synthesis and the induction of the misfolded protein response. Misfolded protein responses are also upregulated by other bacteria such as E. coli (18), P. aeruginosa (41), and M. tuberculosis (47) in response to exposure to copper. In gram-positive bacteria, the Clp proteolytic complex plays an indispensable role in cellular protein quality control by refolding or degrading damaged proteins (11). However, as well as being involved in protein quality control, the CtsR regulon could also control the proteome in response to environmental change through the degradation of key transcriptional regulators. Previous studies suggest that the S. aureus ClpP protease (24) and ClpC ATPase (4) may be involved in regulating copper homeostasis, as a number of genes differentially regulated by copper are also regulated by ClpP and ClpC, such as the copA, copZ, ctsR, clpB, clpC ahpC, sodM, sodA, and trxA genes. The CtsR regulon may be induced by copper due to the presence of misfolded proteins, or the modulators of CtsR repressor activity may sense copper directly. In B. subtilis, the CtsR transcriptional regulator is targeted by MscA and MscB proteins for degradation by the ClpCP protease in the presence of misfolded proteins, hence modulating CtsR action (19). The S. aureus mcsA homologue contains a MXCXXC metal binding motif which may bind, and hence sense, excess copper, thus modulating CtsR activity and therefore inducing a copper response. This possibility needs to be investigated further, particularly as the role of MscA and MscB has not been determined in S. aureus to date.
Our microarray data also show that as well as there being some similarities in the copper response in S. aureus and other bacterial species, there are also significant differences, suggesting that S. aureus has a unique adaptive response to copper. In E. coli and P. aeruginosa, iron homeostasis is significantly affected by copper exposure, as a number of iron transporters and siderophore biosynthetic genes are induced by copper (18, 41). Furthermore, the iron-dependent global regulator fur is also induced by copper in E. coli, P. aeruginosa, and M. tuberculosis, demonstrating a link between copper and iron homeostasis in these organisms. However, our transcriptional profiling shows that while the S. aureus copper regulon contains a number of genes that are also regulated by iron, these are genes involved mainly in the oxidative stress response. In S. aureus, there is no induction of iron uptake transporters, siderophore biosynthetic genes, or fur in response to excess copper, suggesting that copper and iron homeostasis may not be linked in S. aureus. Furthermore, in S. aureus, no homologues of copper-dependent transcriptional regulators are induced in response to high levels of copper. This is in comparison to transcriptional profiling of M. tuberculosis and P. aeruginosa, where high levels of copper induce the expression of the copper-responsive regulators CsoR and CueR and the CopRS two-component regulator system, respectively (41, 47), suggesting that there may be novel mechanisms for induction of the copper response in S. aureus.
Oxidative stress has been implicated in copper toxicity in bacteria such as P. aeruginosa and M. tuberculosis, as transcriptional profiling studies have shown that oxidative stress resistance genes are induced in response to copper stress (41, 47). However, studies in E. coli have shown that copper toxicity is not caused by DNA damage through oxidative stress but occurs mainly through inactivation of the iron-sulfur clusters of the dehydratase enzymes, leading to defective amino acid biosynthetic pathways (22). It is likely that copper-mediated inactivation of biosynthetic enzymes does occur in S. aureus, as SH1000 copper sensitivity is increased in CRPMI, which is an extremely nutrient-poor medium compared to other richer media such as DM and TSB (37). However, both our microarray and phenotypic studies demonstrate that oxidative stress resistance is important for S. aureus exposed to high-copper conditions.
In the absence of KatA and AhpC, S. aureus demonstrated an increased sensitivity to copper, whereas the absence of SodA and SodM decreased sensitivity, suggesting that superoxide dismutase activity is actually contributing to copper toxicity. This can be explained by the Haber-Weiss reaction, as copper can act as a catalyst with H2O2 to create toxic hydroxyl radicals, as follows: Cu2+ + O2− → Cu1+ + O2 (reaction 1); Cu1+ + H2O2 → Cu2+ + O2 + OH− + OH (reaction 2). Therefore, in the absence of all cellular SOD activity, there is less H2O2 to react with the copper. As a result, reaction 2 does not occur, hence showing increased resistance to copper. Therefore, H2O2 scavenging is an important S. aureus copper resistance mechanism. This agrees with our previous results which demonstrated that the copper efflux copA mutant shows increased sensitivity to H2O2 (37).
Previous studies demonstrated that copper toxicity in Saccharomyces cerevisiae was not due entirely to excess intracellular copper but was due to surface-produced Cu1+ (34), as the increased copper sensitivity observed due to the deletion of a surface-associated oxidase, Fet3p, was suppressed by the codeletion of the surface ferric/cupric reductase but not by the deletion of the major copper uptake transporters. Our results showing the protective effects of extracellular ROS quenchers and the importance of H2O2 suggest that copper toxicity is also occurring at the cell surface in S. aureus. It is not clear how surface Cu1+ is produced on the staphylococcal cell surface, but the oxidative stress enzymes do appear to be required for resistance to this surface toxicity. This would seem unlikely, as the oxidative stress enzymes are supposed to be intracellular; however, they have been found associated with cell surface extracts in S. aureus (35), and therefore, our data suggest that this localization may not just be experimental artifact but may have physiological relevance. This evidence of extracellular copper toxicity may also help explain why copper surfaces are so effective at preventing S. aureus survival (28).
As well as inducing stress responses, excess copper also leads to the repression of two major global virulence gene regulators, sae and agr. Biofilm formation is an extremely important S. aureus virulence factor. Copper surface films on catheters show antibacterial activity against S. aureus biofilms (23), but it was not known why. Therefore, our observation that copper represses sae and agr, which results in the repression of proteins involved in biofilm formation, such as Eap, has important implications for the effect of copper on virulence and the antimicrobial uses of copper.
In summary, our data suggest that copper toxicity in S. aureus is caused through copper catalyzing the intracellular and extracellular generation of oxidative stress through the Haber-Weiss reaction and by affecting the function/folding of proteins. Hence, excess environmental copper leads to the stimulation of a global response, which includes the induction of the copper, oxidative stress, and misfolded protein responses and the repression of two important global regulators, Agr and Sae, both of which are essential for S. aureus virulence (Fig. 2).
Supplementary Material
Acknowledgments
We thank Simon Foster for providing the SH1000 ahpC, katA, sodM, sodA, and sodM sodA mutants. The DNA microarrays were obtained through NIAID's Pathogen Functional Genomics Resource Center, Division of Microbiology and Infectious Diseases, NIH. We thank Alan Cockayne and Peter Williams for their helpful comments on the manuscript.
This work was partially supported by a grant from NIH to R.K.J.
Footnotes
Published ahead of print on 30 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arredondo, M., and M. T. Nunez. 2005. Iron and copper metabolism. Mol. Aspects Med. 26:313-327. [DOI] [PubMed] [Google Scholar]
- 2.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 3.Catalani, S., G. De Palma, A. Mangili, and P. Apostoli. 2008. Metallic elements in lung tissues: results of a meta-analysis. Acta Biomed. 79(Suppl. 1):52-63. [PubMed] [Google Scholar]
- 4.Chatterjee, I., S. Schmitt, C. F. Batzilla, S. Engelmann, A. Keller, M. W. Ring, R. Kautenburger, W. Ziebuhr, M. Hecker, and K. T. Preissner. 2009. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9:1152-1176. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U. S. A. 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements, M. O., S. P. Watson, and S. J. Foster. 1999. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181:3898-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove, K., G. Coutts, I. M. Jonsson, A. Tarkowski, J. F. Kokai-Kun, J. J. Mond, and S. J. Foster. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 10.Francis, M. S., and C. J. Thomas. 1997. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb. Pathog. 22:67-78. [DOI] [PubMed] [Google Scholar]
- 11.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, R. J., and F. D. Lowy. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46(Suppl. 5):S350-S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U. S. A. 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiron, A., E. Borezee-Durant, J. Piard, and V. Juillard. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, M., A. Cockayne, and J. A. Morrissey. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76:1756-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karavolos, M. H., M. J. Horsburgh, E. Ingham, and S. J. Foster. 2003. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology 149:2749-2758. [DOI] [PubMed] [Google Scholar]
- 18.Kershaw, C. J., N. L. Brown, C. Constantinidou, M. D. Patel, and J. L. Hobman. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187-1198. [DOI] [PubMed] [Google Scholar]
- 19.Kirstein, J., and K. Turgay. 2005. A new tyrosine phosphorylation mechanism involved in signal transduction in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 9:182-188. [DOI] [PubMed] [Google Scholar]
- 20.Liu, T., A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini, and D. P. Giedroc. 2007. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3:60-68. [DOI] [PubMed] [Google Scholar]
- 21.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 22.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean, R. J., A. A. Hussain, M. Sayer, P. J. Vincent, D. J. Hughes, and T. J. Smith. 1993. Antibacterial activity of multilayer silver-copper surface films on catheter material. Can. J. Microbiol. 39:895-899. [DOI] [PubMed] [Google Scholar]
- 24.Michel, A., F. Agerer, C. R. Hauck, M. Herrmann, J. Ullrich, J. Hacker, and K. Ohlsen. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morikawa, K., R. L. Ohniwa, J. Kim, A. Maruyama, T. Ohta, and K. Takeyasu. 2006. Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells 11:409-423. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey, J. A., A. Cockayne, J. Hammacott, K. Bishop, A. Denman-Johnson, P. J. Hill, and P. Williams. 2002. Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus. Infect. Immun. 70:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289-297. [DOI] [PubMed] [Google Scholar]
- 29.Portmann, R., K. R. Poulsen, R. Wimmer, and M. Solioz. 2006. CopY-like copper inducible repressors are putative ‘winged helix’ proteins. Biometals 19:61-70. [DOI] [PubMed] [Google Scholar]
- 30.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai, T., and K. Kataoka. 2007. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem. Rec. 7:220-229. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwan, W. R., P. Warrener, E. Keunz, C. K. Stover, and K. R. Folger. 2005. Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. Int. J. Med. Microbiol. 295:237-242. [DOI] [PubMed] [Google Scholar]
- 34.Shi, X., C. Stoj, A. Romeo, D. J. Kosman, and Z. Zhu. 2003. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 278:50309-50315. [DOI] [PubMed] [Google Scholar]
- 35.Sibbald, M. J., A. K. Ziebandt, S. Engelmann, M. Hecker, A. de Jong, H. J. Harmsen, G. C. Raangs, I. Stokroos, J. P. Arends, J. Y. Dubois, and J. M. van Dijl. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70:755-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sitthisak, S., K. Howieson, C. Amezola, and R. K. Jayaswal. 2005. Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl. Environ. Microbiol. 71:5650-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitthisak, S., L. Knutsson, J. W. Webb, and R. K. Jayaswal. 2007. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology 153:4274-4283. [DOI] [PubMed] [Google Scholar]
- 38.Smaldone, G. T., and J. D. Helmann. 2007. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 153:4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183-195. [DOI] [PubMed] [Google Scholar]
- 40.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teitzel, G. M., A. Geddie, S. K. De Long, M. J. Kirisits, M. Whiteley, and M. R. Parsek. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188:7242-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Townsend, D. E., and B. J. Wilkinson. 1992. Proline transport in Staphylococcus aureus: a high-affinity system and a low-affinity system involved in osmoregulation. J. Bacteriol. 174:2702-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veldhuis, N. A., A. P. Gaeth, R. B. Pearson, K. Gabriel, and J. Camakaris. 2009. The multi-layered regulation of copper translocating P-type ATPases. Biometals 22:177-190. [DOI] [PubMed] [Google Scholar]
- 45.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Said-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907-3919. [DOI] [PubMed] [Google Scholar]
- 46.Voyich, J. M., C. Vuong, M. Dewald, T. K. Nygaard, S. Kocianova, S. Griffith, J. Jones, C. Iverson, D. E. Sturdevant, K. R. Braughton, A. R. Whitney, M. Otto, and F. R. Deleo. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 6:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, S. K., E. A. Hoye, and A. M. Talaat. 2008. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190:2939-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.