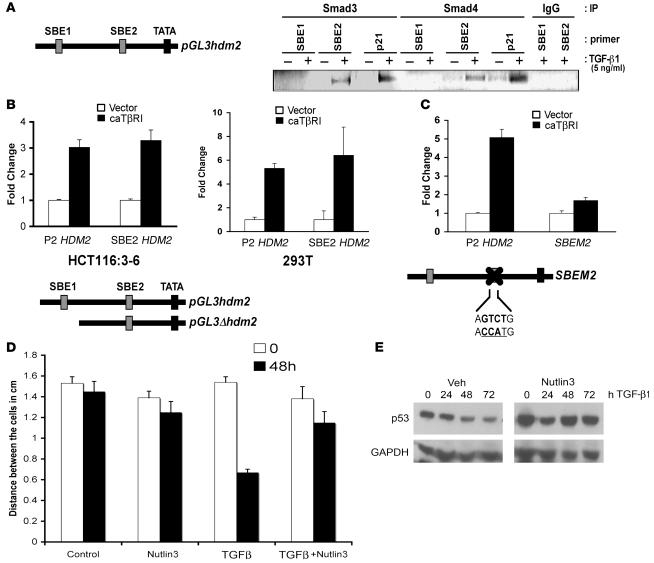
Figure 5. Binding of Smad3 and Smad4 to the HDM2 SBE2 in vivo.
(A) HCT116:3-6 cells were treated with vehicle or TGF-β1 (10 ng/ml) for 48 hours. Smad3, Smad4, or IgG isotype control antibodies were used to immunoprecipitate Smads binding to the HDM2 promoter. Oligo sets were used to PCR amplify the SBE1, SBE2, or the p21 promoter SBE (a positive control for Smad3/4 binding) and resolved on an agarose gel. (B) HCT116:3-6 and 293T cells were transfected with caTβRI and either the full-length P2 HDM2-promoter driving luciferase or a deletion construct missing SBE1. Reporter activity was determined relative to renilla to generate relative activity. Fold induction was determined relative to vehicle control, and SD was calculated relative to the mean. (C) Cells were transfected with caTβRI and the P2 HDM2 promoter driving luciferase or a mutant whereby the SBE2 site was mutated. A renilla expression construct was used as an internal control for normalization. SEM was calculated as in B. (D) Wound healing assay using HCT116:3-6 cells, scored in a double-blind assay. Bar graph represents the distance between confluent cells after stimulation with TGF-β1 (10 ng/ml), Nutlin3 (10 μM), or TGF-β1 (10 ng/ml) and Nutlin3 (10 μM). (E) Western blot analysis of p53 in HCT116:3-6 cells treated with TGF-β1 (10 ng/ml) with or without Nutlin3 (10 μM) over a 72-hour time course. SD was calculated relative to the mean.