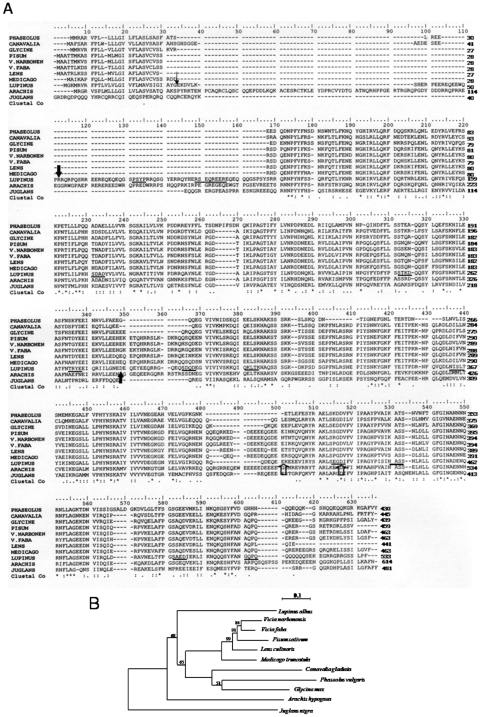
Figure 1. Comparison of β-conglutin precursor with the vicilins from other legume species.
(A) Multiple sequence alignment of predicted amino acid sequences of Lupinus albus β-conglutin precursor with other legume vicilins. In the β-conglutin sequence, the small arrowhead designates the putative cleavage of the signal peptide; larger closed arrowheads denote the N- and C- terminals of blad; larger open arrowheads denote the beginning and the end of the UIM; potential N-glycosylation and phosphorylation sites are underlined by thick and thin lines, respectively. The asterisks indicate that the residues in column are identical in all sequences in the alignment; colons indicate conservative substitutions; full stops mean that semi-conserved substitutions are observed. Hyphens are inserted to optimize the alignments. LUPINUS- Lupinus albus; V.FABA- Vicia faba; LENS- Lens culinaris; PISUM- Pisum sativum; V.NARBORENSIS- Vicia narborensis, GLYCINE- Glycine max; PHASEOLUS- Phaseolus vulgaris; CANAVALIA- Canavalia gladiata; MEDICAGO- Medicago truncatula; JUGLANS- Juglans nigra; ARACHIS- Arachis hypogaea. (B) Phylogram of vicilin-like proteins with high homology to β-conglutin from Lupinus albus. The following proteins were included in the phylogenetic analyses: Vicia faba (vicilin precursor, CAA68525), Lens culinaris (allergen Len c, CAD87731), Pisum sativum (vicilin, CAA32239), Vicia narbonensis (vicilin precursor, CAA96514), Glycine max (alpha' subunit of beta-conglycinin, BAE02726), Phaseolus vulgaris (phaseolin, AAC04316), Canavalia gladiata (canavalin, CAA33172), Medicago truncatula (cupin, ABD28364), Juglans nigra (vicilin, AAM54366) and Arachis hypogaea (allergen Ara h 1, P43237).