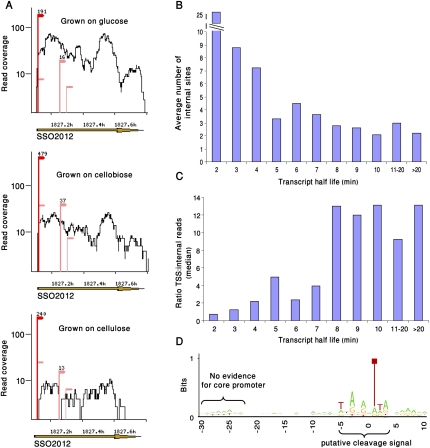
Figure 4.
Sequencing the 5′ ends of RNA degradation products. (A) Internal sites in RNAs. Shown are the transcriptome data on gene SSO2012. (Red) TSS; (pink) internal sites. The three different sequencing samples are presented to show the reproducibility of internal sites between samples. Numbers above dominant sites represent the number of reads supporting the site. (B) Negative correlation between the observed number of internal sites in our data and transcript half-life as measured in a global survey of S. solfataricus RNA stability (Andersson et al. 2006). Transcripts with shorter half-lives tend to contain a higher number of internal sites (Spearman correlation, ρ = −0.9). (C) Ratio between the number of reads in TSS and number of internal reads is positively correlated with transcript half-life (Spearman correlation, ρ = 0.9). (D) A sequence motif at the position of internal sites. Nucleotide frequency at internal sites (n = 1240) was plotted using the WebLogo tool (Crooks et al. 2004). Position 0 represents the first sequenced nucleotide, that is, the first nucleotide 3′ to the RNA cleavage position. The core promoter is largely absent from the region upstream to this motif.