Figure 1.
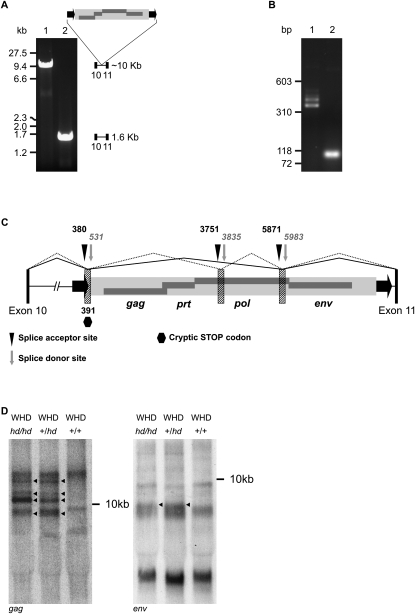
Insertion of a retrovirus into Cntrob. (A) Long-range PCR spanning exons 10 and 11 shows a ∼10-kb insertion in hd/hd homozygotes (lane 1), whereas the SHR control PCR product (lane 2) has the expected size (1578 bp). The Cntrob gene contains insertion of an endogenous retrovirus (ERV) in the sense orientation, containing all typical retroviral genes (gag, pol, prt, and env, dark gray rectangles) flanked by long terminal repeats (LTRs, black arrows). Exons 10 and 11 are represented by black rectangles. (B) Aberrant splicing of Cntrob due to the retroviral insertion. RT-PCR of total RNA from testes spanning exons 10 and 11 of Cntrob. Lane 1 shows multiple PCR products in a size range of 400–500 bps in mutant (hd/hd) rats. Lane 2 shows the expected 98-bp PCR product in SHR controls (+/+). (C) Cntrob-ERV chimeric transcripts identified by RT-PCR. Splice signals are designated as vertical, black triangles (splice acceptor) or as vertical, gray arrows (splice donor). Up to three short segments of the retrovirus (striped rectangles [not to scale]) incorporate into the mature Cntrob mRNA. Exons 10 and 11 of Cntrob are depicted as black bars (not to scale). Two out of five different splicing variants are shown (solid and dotted lines). STOP codon is marked as a hexagon. (D) Northern blotting of total RNA from testes of WHDhd/hd, WHD+/hd, and WHD+/+. The probe corresponded to either a gag (left panel) or an env (right panel) fragment of the Cntrob-ERV. A <9-kb transcript is detectable in all the samples. The multiple hd-specific bands are marked by arrows.