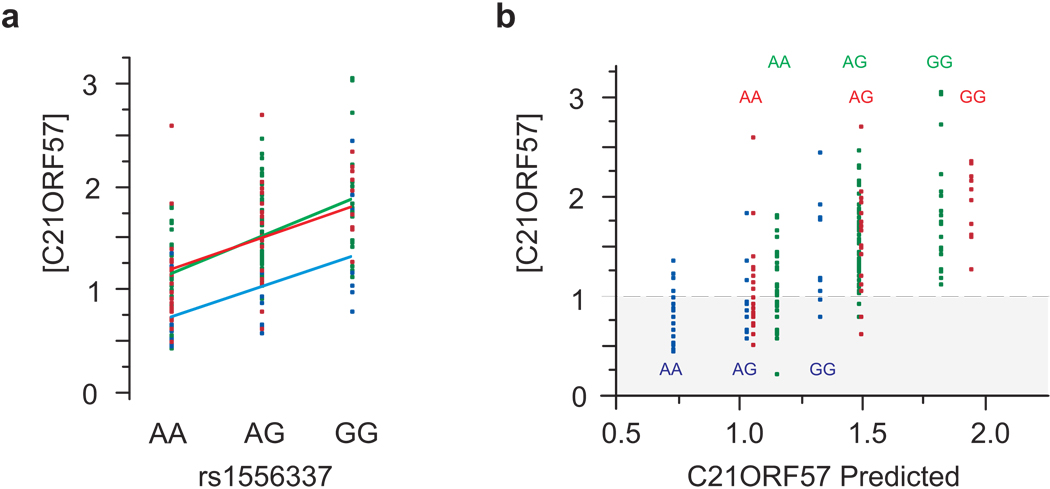
Figure 6. The relationship between genotype, expression, and phenotype.
(a) A typical example of a transcript (encoding C21ORF57, a putative metallo-proteinase) that shows both a significant difference between locations (P < 10−5) and a cis-eSNP association, with rs1556337 (P < 10−13) but no interaction effect in an additive model on the log scale. Expression is lower in Boutroch (blue points and line), while genotype has a consistent effect across all three locations (Ighrem, red; Agadir, green). (b) The Actual vs Predicted plot separates the genotypes by location for clarity. Suppose that a disease or phenotype is only seen in individuals with transcript abundance less than 1.0 (on a relative log2 scale), indicated by the gray area. Then in Agadir and Ighrem (green and red respectively) almost all affected are AA homozygotes, whereas in Boutroch (blue) heterozygotes and some GG homozygotes are also affected. There is thus a G×E interaction for the phenotype in the absence of a G×E interaction for transcription, because the environment shifts more individuals into the susceptible zone. Similar arguments would apply for phenotypes with high expression values, and for graded rather than threshold-dependent traits.