Table 1. Benchmarking results for ODM, NRM, MODM and LOLCAT Method.
System Parameters Runtimes(s) 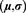
| ||||||
| ID | # Rxns. | # Species | ODM | NRM | MODM | LOLCAT |
| A | 2040 | 236 | 8.45, 0.04 | 25.52, 5.75 | 25.08, 0.12 | 3.06, 0.14 |
| B | 11492 | 5092 | 61.08, 0.17 | 162.68, 6.15 | 64.21, 0.22 | 1.68, 0.04 |
| C | 35003 | 11402 | 152.18, 0.64 | 374.73, 4.63 | 166.06, 0.46 | 3.73, 0.06 |
| D | 84301 | 15087 | 374.66, 2.30 | 720.35, 62.32 | 390.85, 1.41 | 4.73, 0.13 |
| E | 162150 | 14766 | 640.72, 3.01 | 964.49, 99.97 | 623.39, 1.58 | 1.88, 0.04 |
| F | 292190 | 15287 |

|

|
2527.93, 61.09 | 9.30, 0.09 |
Six versions of the Yeast Pheromone Model [10] were benchmarked. Slowdown factors are mean time normalized against the performance of LOLCAT Method for each model. The “ ” in the last entry for ODM and NRM indicates that those simulations could not be run because the dependency graph consumed more than the RAM and swap space, roughly 20 GB, of the host machine. The varying structure of the simulated system may account for the non-uniform scaling of runtimes.
” in the last entry for ODM and NRM indicates that those simulations could not be run because the dependency graph consumed more than the RAM and swap space, roughly 20 GB, of the host machine. The varying structure of the simulated system may account for the non-uniform scaling of runtimes.
