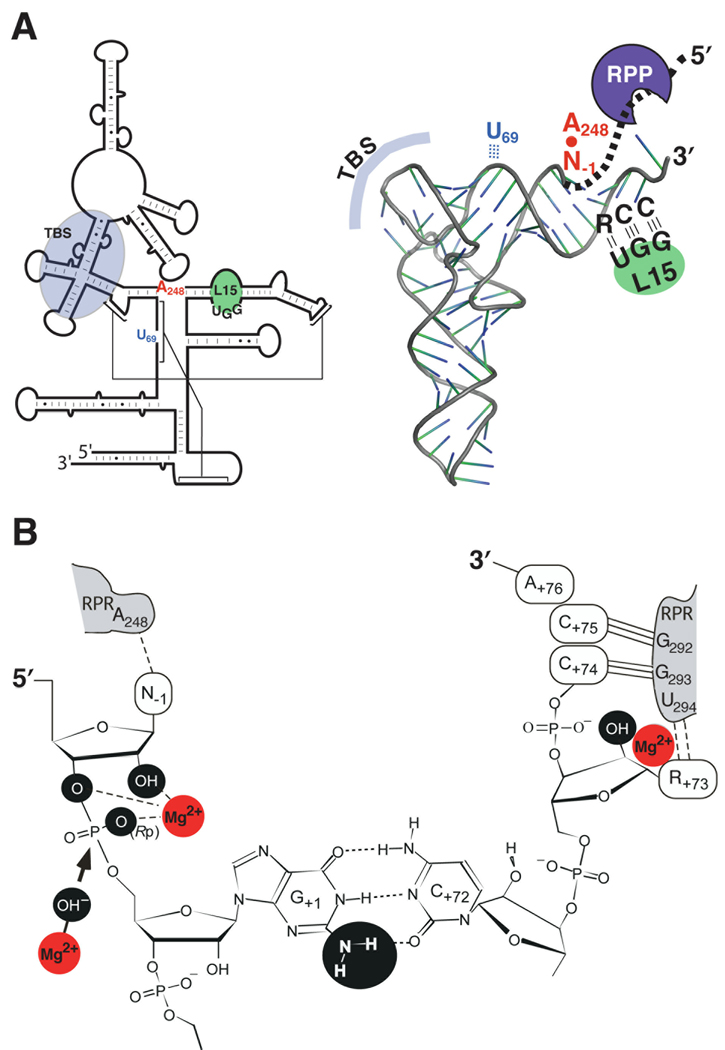
Figure 2.
Substrate recognition and catalysis by bacterial RNase P (see section 3 for details). (A) Secondary structures of the E. coli RPR (left) and a pre-tRNA (right) illustrating the domains required for interactions during RNase P catalysis. The interaction of the leader sequence in the pre-tRNA (bold, dashed line) with the RPP is also indicated. TBS, T stem-loop-binding site. (B) A close-up view showing one model of the canonical RNase P cleavage site. Chemical groups (black spheres) and Mg2+ (red spheres) suggested to contribute to catalysis are marked. Also depicted is a Mg2+-activated hydroxide nucleophile (arrow), which attacks the phosphorous atom in the scissile bond.