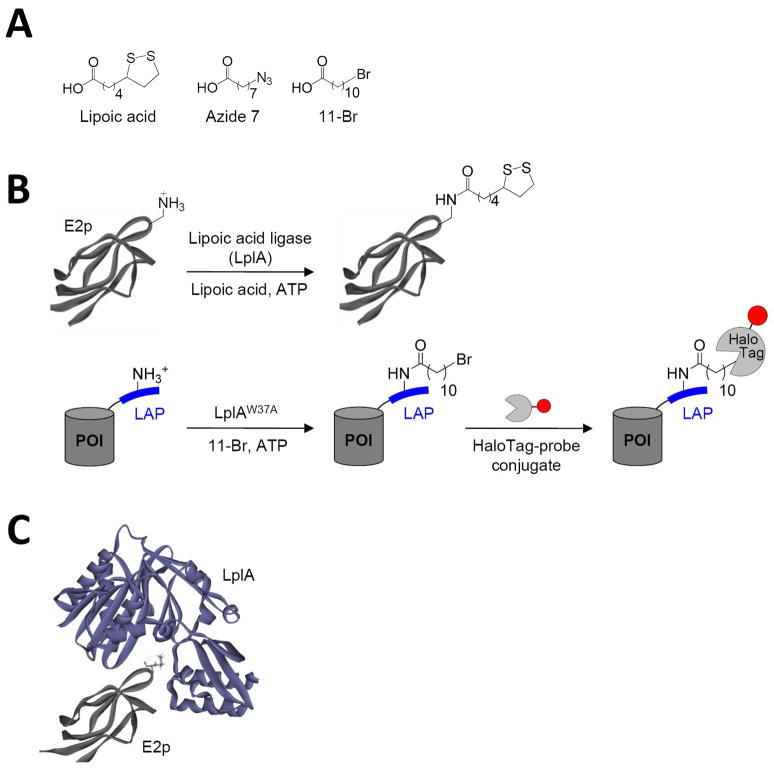
Figure 1.
LplA-catalyzed protein and peptide labeling reactions. (A) Natural (lipoic acid) and unnatural (Azide 7 and 11-Br) small-molecule substrates of LplA and its W37 mutant. (B) Natural and engineered ligation reactions. Top: LplA-catalyzed lipoylation of the 9 kD E2p domain of E. coli pyruvate dehydrogenase (structure from PDB 1QJO). Bottom: LplA-catalyzed 11-Br ligation onto an engineered LAP (“LplA Acceptor Peptide”), which is genetically fused to any protein of interest (POI). Ligated alkyl bromide can be specifically and covalently modified by HaloTag-fluorophore conjugates.31 The red circle represents any probe. (C) Model for interaction between LplA (purple, from PDB 1X2H)33 and E2p (grey, from PBD 1QJO).34 The lipoylation site on E2p, Lys41, is rendered in stick.