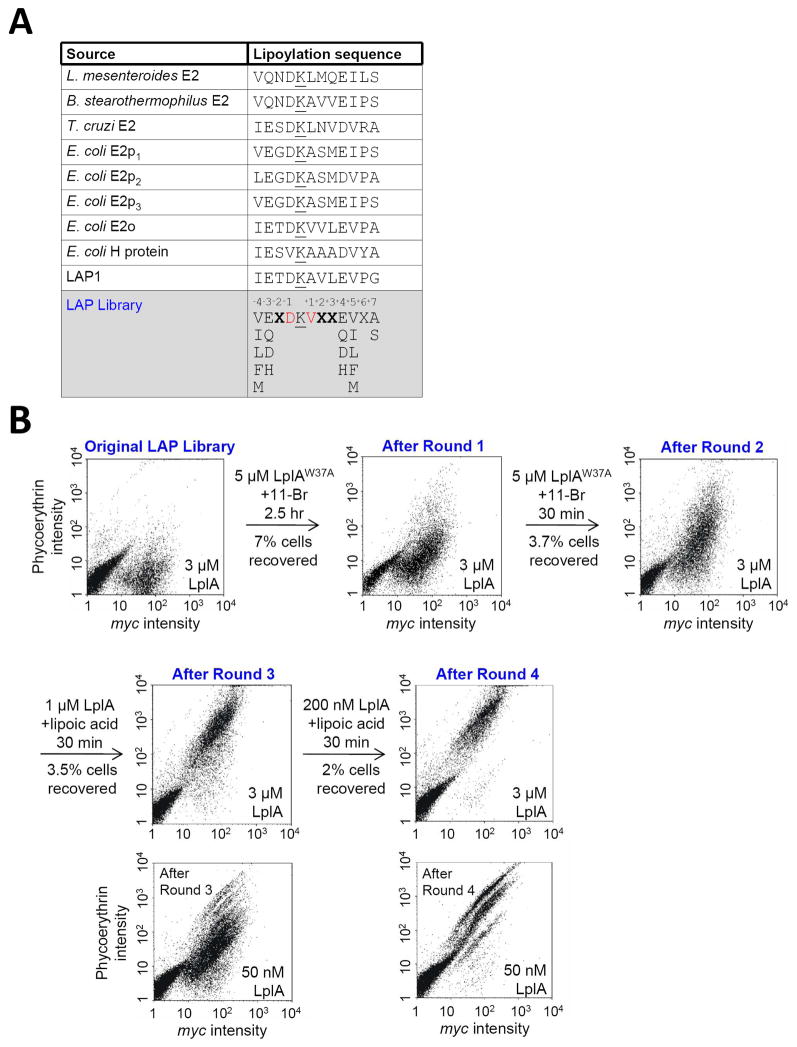
Figure 3.
Library design and selection results. (A) Table showing sequences of natural LplA protein substrates, our previous rationally-designed LAP1,13 and our LAP library. Lysine modification sites are underlined. For the LAP library, we fixed positions −4 and +5 as hydrophobic amino acids (Val, Ileu, Leu, Phe, Met), positions −3 and +4 as polar amino acids (Glu, Asp, Gln, His), and position +7 as Ser or Ala. Red-colored positions are partially randomized (39% Asp or 49% Val). X represents any amino acid. (B) Results of four rounds of selection. Selection conditions, including small-molecule substrates used for labeling, are given above each arrow. To analyze amplified yeast pools following each round of selection, uniform lipoylation conditions were used (given in the lower right of each scatter plot). Yeast pools from rounds 3 and 4 were additionally analyzed under milder conditions, with 50 nM LplA.