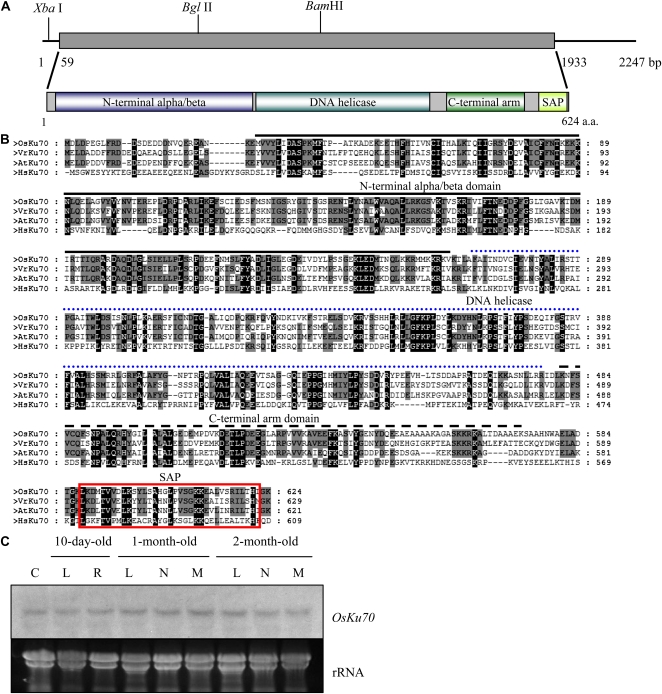
Figure 1.
Sequence analysis of rice OsKu70. A, Schematic structure of OsKu70 cDNA and its predicted protein. The solid bar represents the coding region, while the solid line indicates 5′ and 3′ untranslated regions. The N-terminal α/β-domain, DNA helicase motif, C-terminal arm domain, and SAP DNA-binding domain are indicated. a.a., Amino acids. B, Comparison of the deduced amino acid sequences of Ku70 homologs from rice, mung bean, Arabidopsis, and human. Amino acid residues conserved in at least three of the four sequences are shaded. Amino acids identical in all four proteins are highlighted in black. The N-terminal α/β-domain, DNA helicase motif, C-terminal arm domain, and SAP DNA-binding domain are indicated by a solid line, dotted line, broken line, and box, respectively. C, RNA gel-blot analysis of OsKu70 mRNA in wild-type rice plants. Total RNA (40 μg per lane) was obtained from various organs at different developmental stages as indicated, separated on an agarose gel, and blotted onto a nylon membrane. The membrane was hybridized to a 32P-labeled OsKu70 cDNA probe under high-stringency conditions. rRNA is shown as a loading control. C, Callus; L, leaf; R, root; N, internode; M, meristem. [See online article for color version of this figure.]