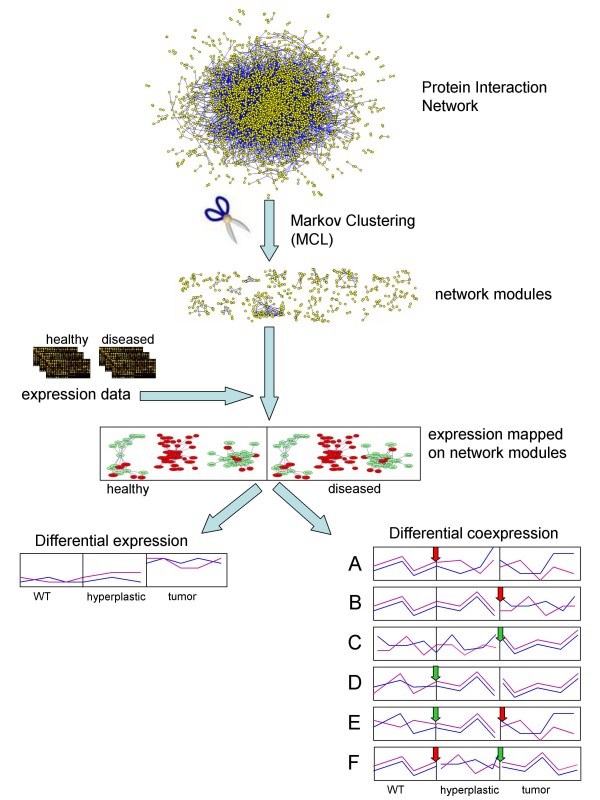
Figure 1.
Outline of the analysis. The mouse IntNetDB Protein Interaction Network (PIN) was decomposed into into highly interconnected subgraphs, or modules, with Markov graph clustering [23]. Gene expression values from three stages of mammary cancer (healthy, hyperplastic and tumor [14]) were mapped on the proteins in network modules. Next, the modules were tested for differential expression and differential coexpression between the disease stages. Plots A-F show schematic patterns of formation and vanishing of the coexpression in the module. (A-B) Modules coexpressed in healthy tissues lose their coordination in the diseased tissues. Such modules may represent processes that are disorganized in cancer. (C-D) Coexpression is not present in healthy tissue, but appears in the diseased ones. Such behavior may characterize processes that are invoked in cancer. (E-F) Correlation appears (or is lost) only in the hyperplastic tissue, indicating processes transiently active (or disrupted) in early stage of disease. The red arrow indicates loss of the correlation, green arrow marks a gain of the correlation by the module. The PIN is drawn with Pajek software [59].